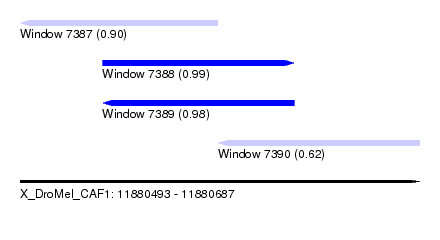
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,880,493 – 11,880,687 |
| Length | 194 |
| Max. P | 0.989161 |

| Location | 11,880,493 – 11,880,589 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 96.46 |
| Mean single sequence MFE | -23.32 |
| Consensus MFE | -20.82 |
| Energy contribution | -20.70 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11880493 96 - 22224390 UGCACACACAUUCAGACGCAUAUGUAUGGAUCAGUGUGCAUUUGUGUGUGUGCUUCUGGCGACGCAUUAUUUUGUAUUUUAAUAAUUCAAAUAUCA .(((((((((..((((.((((((..........)))))).)))))))))))))..........(((......)))..................... ( -23.60) >DroSec_CAF1 12484 96 - 1 UGCACACACAUUCAGUCGCAUAUGUAUGGAUCAGUGUGCAUUUGUGUGUGUGCUUCUGGCGACGCAUUAUUUUGUAUUUUAAUAAUUCAAAUAUCA .((((((((((..(((.((((((..........))))))))).))))))))))..........(((......)))..................... ( -22.50) >DroSim_CAF1 14821 96 - 1 UGCACACACAUUCAGUCGCAUAUGUAUGGAUCAGUGUGCAUUUGUGUGUGUGCUUCUGGCGACGCAUUAUUUUGUAUUUUAAUAAUUCAAAUAUCA .((((((((((..(((.((((((..........))))))))).))))))))))..........(((......)))..................... ( -22.50) >DroEre_CAF1 14620 96 - 1 UGCACACACAUUCAGUCGCCAGUGUGUGGAUCAGCGUGCAUUUGUGUGUGUGCUUCUGGCGACGCAUUAUUUUGUAUUUUAAUAAUUCAAAUAUCA ..............((((((((..........((((..(((....)))..)))).))))))))(((......)))..................... ( -24.10) >DroYak_CAF1 16135 96 - 1 UGCACACACGUUCAGUCGCAUGUGUAUGGAUCAGUGUGCAUUUGUGUGUGCGCUUCUGGCGACGCAUUAUUUUGUAUUUUAAUAAUUCAAAUAUCA (((((((..(((((...((....)).)))))..)))))))...((((((.(((.....)))))))))...((((.(((.....))).))))..... ( -23.90) >consensus UGCACACACAUUCAGUCGCAUAUGUAUGGAUCAGUGUGCAUUUGUGUGUGUGCUUCUGGCGACGCAUUAUUUUGUAUUUUAAUAAUUCAAAUAUCA (((((((..(((((...((....)).)))))..)))))))...((((((.(((.....)))))))))...((((.(((.....))).))))..... (-20.82 = -20.70 + -0.12)

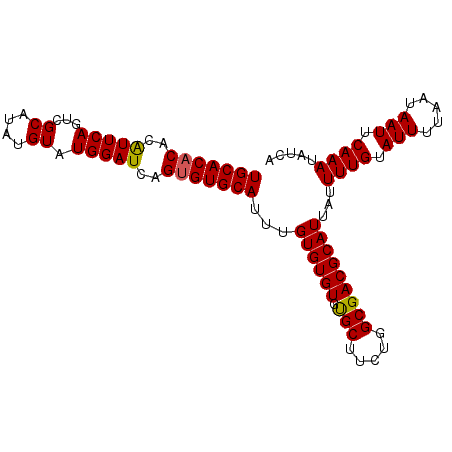
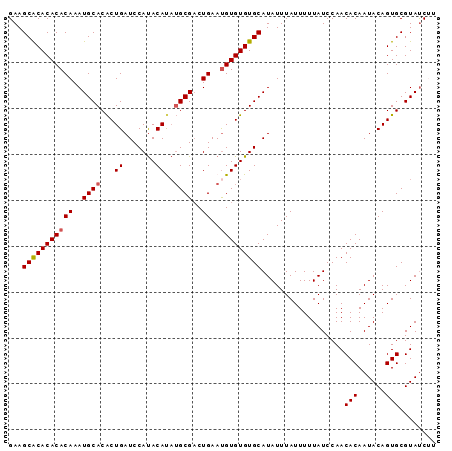
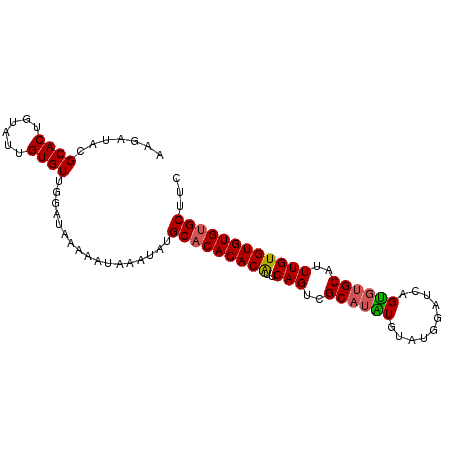
| Location | 11,880,533 – 11,880,626 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 94.73 |
| Mean single sequence MFE | -19.22 |
| Consensus MFE | -17.22 |
| Energy contribution | -17.46 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11880533 93 + 22224390 GAAGCACACACACAAAUGCACACUGAUCCAUACAUAUGCGUCUGAAUGUGUGUGCAUAUUUAUUUUUAUCCAACACAAUACAGUGCGUACCGU ...(((((((((((.(((((...((.......))..))))).))..)))))))))..................(((......)))........ ( -20.60) >DroSec_CAF1 12524 93 + 1 GAAGCACACACACAAAUGCACACUGAUCCAUACAUAUGCGACUGAAUGUGUGUGCAUAUUUAUUUUUAUCCAACACAAUACAGUGCGUAUCUU ...(((((((((((..((((...((.......))..))))..))..)))))))))..................(((......)))........ ( -18.30) >DroSim_CAF1 14861 93 + 1 GAAGCACACACACAAAUGCACACUGAUCCAUACAUAUGCGACUGAAUGUGUGUGCAUAUUUAUUUUUAUCCAACACAAUACAGUGCGUAUCUA ...(((((((((((..((((...((.......))..))))..))..)))))))))..................(((......)))........ ( -18.30) >DroEre_CAF1 14660 93 + 1 GAAGCACACACACAAAUGCACGCUGAUCCACACACUGGCGACUGAAUGUGUGUGCAUAUUUAUUUUUAUCCAACACAAUACAGUGCGUAUCAU ((.(((((((((((..(((.((.((.......)).)))))..))..)))))))))..................(((......)))....)).. ( -19.20) >DroYak_CAF1 16175 93 + 1 GAAGCGCACACACAAAUGCACACUGAUCCAUACACAUGCGACUGAACGUGUGUGCAUAUUUAUUUUUAUCCAACACAAUACAGUGCGUAUCUU ((.((((((......((((((((((.......))...(((......))))))))))).........(((........)))..)))))).)).. ( -19.70) >consensus GAAGCACACACACAAAUGCACACUGAUCCAUACAUAUGCGACUGAAUGUGUGUGCAUAUUUAUUUUUAUCCAACACAAUACAGUGCGUAUCUU ...(((((((((((..((((...((.......))..))))..))..)))))))))..................(((......)))........ (-17.22 = -17.46 + 0.24)

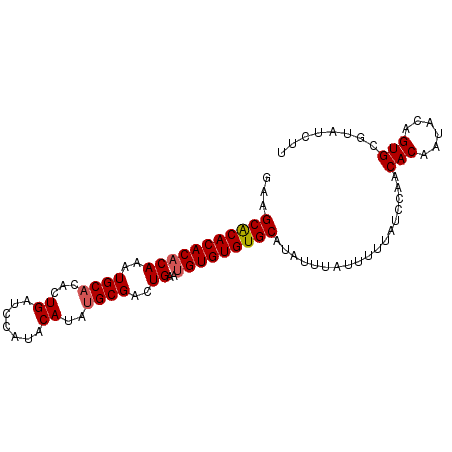
| Location | 11,880,533 – 11,880,626 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 94.73 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -21.94 |
| Energy contribution | -21.98 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11880533 93 - 22224390 ACGGUACGCACUGUAUUGUGUUGGAUAAAAAUAAAUAUGCACACACAUUCAGACGCAUAUGUAUGGAUCAGUGUGCAUUUGUGUGUGUGCUUC ..(((((((.(((...(((((((.(((........))).)).)))))..)))(((((.((((((........)))))).)))))))))))).. ( -29.30) >DroSec_CAF1 12524 93 - 1 AAGAUACGCACUGUAUUGUGUUGGAUAAAAAUAAAUAUGCACACACAUUCAGUCGCAUAUGUAUGGAUCAGUGUGCAUUUGUGUGUGUGCUUC .......((((......)))).................((((((((((..(((.((((((..........))))))))).))))))))))... ( -25.00) >DroSim_CAF1 14861 93 - 1 UAGAUACGCACUGUAUUGUGUUGGAUAAAAAUAAAUAUGCACACACAUUCAGUCGCAUAUGUAUGGAUCAGUGUGCAUUUGUGUGUGUGCUUC .......((((......)))).................((((((((((..(((.((((((..........))))))))).))))))))))... ( -25.00) >DroEre_CAF1 14660 93 - 1 AUGAUACGCACUGUAUUGUGUUGGAUAAAAAUAAAUAUGCACACACAUUCAGUCGCCAGUGUGUGGAUCAGCGUGCAUUUGUGUGUGUGCUUC .((((((((((((...(((((((.(((........))).)).))))).........))))))))..))))(((..(((....)))..)))... ( -25.60) >DroYak_CAF1 16175 93 - 1 AAGAUACGCACUGUAUUGUGUUGGAUAAAAAUAAAUAUGCACACACGUUCAGUCGCAUGUGUAUGGAUCAGUGUGCAUUUGUGUGUGCGCUUC ...((((((((((...(((((((.(((........))).)).)))))..)))).....)))))).....((((..(((....)))..)))).. ( -24.10) >consensus AAGAUACGCACUGUAUUGUGUUGGAUAAAAAUAAAUAUGCACACACAUUCAGUCGCAUAUGUAUGGAUCAGUGUGCAUUUGUGUGUGUGCUUC .......((((......)))).................(((((((((..(((..((((((..........))))))..))))))))))))... (-21.94 = -21.98 + 0.04)

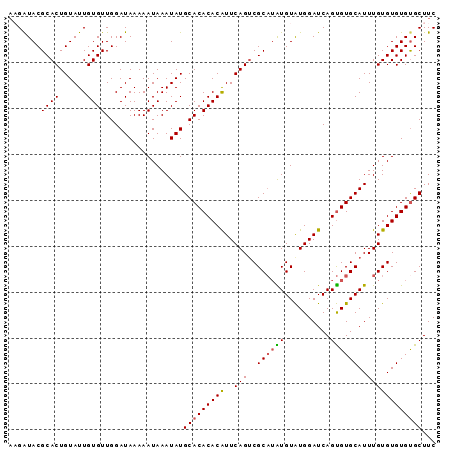
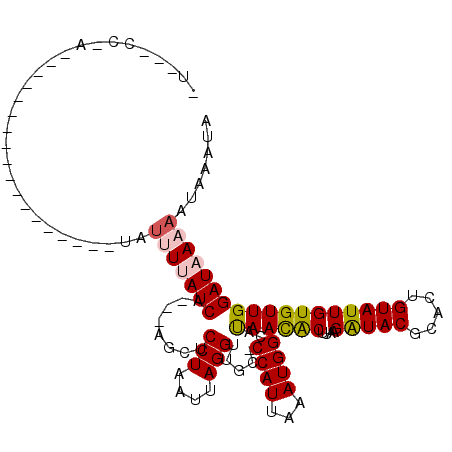
| Location | 11,880,589 – 11,880,687 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.04 |
| Mean single sequence MFE | -19.45 |
| Consensus MFE | -10.36 |
| Energy contribution | -10.56 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11880589 98 - 22224390 UUU--CCUA-U-------------UUGUCUUUUAUCA---AGCUCCUAAUUAGGUUCCCCCAUUAAAUGGCAUAACACUUACGGUACGCACUGUAUUGUGUUGGAUAAAAAUAAAUA ...--..((-(-------------((((.(((((((.---.(((..((((..((...))..))))...))).((((((.((((((....))))))..)))))))))))))))))))) ( -21.50) >DroSec_CAF1 12580 85 - 1 ----------------------------GUUUUAUCC---AGCUCCUAAUUAGGUUGC-CCAUUAAAUGGCAUAACACUUAAGAUACGCACUGUAUUGUGUUGGAUAAAAAUAAAUA ----------------------------.((((((((---(((......(((((((((-(........))))....))))))(((((.....)))))..)))))))))))....... ( -22.10) >DroSim_CAF1 14917 85 - 1 ----------------------------GUUUUAUCA---AGCUCCUAAUUAGGUUGC-CCAUUAAAUGGCAUAACACUUUAGAUACGCACUGUAUUGUGUUGGAUAAAAAUAAAUA ----------------------------.(((((((.---....(((....))).(((-(........))))((((((....(((((.....))))))))))))))))))....... ( -16.30) >DroEre_CAF1 14716 116 - 1 AUUACCCCAGCAUGAAAUAAUACCUCAUACCUCAUCGCAUAGCUCCUAAUUAGGUUCC-CCAUCAAAUGGCCCAAUGCUUAUGAUACGCACUGUAUUGUGUUGGAUAAAAAUAAAUA ......(((((((.....(((((........((((.((((....(((....)))....-((((...))))....))))..))))........))))))))))))............. ( -17.29) >DroYak_CAF1 16231 108 - 1 AUAUUCCCAGCCUGUAA-----CUGCAUAUCUCAUCG---AGAUCCUAAUUAGGUUGC-CCAUCAAAUGGCCCAAUACUUAAGAUACGCACUGUAUUGUGUUGGAUAAAAAUAAAUA .......(((((((...-----......(((((...)---))))......))))))).-((((...)))).(((((((....(((((.....))))))))))))............. ( -20.06) >consensus _U___CC_A__________________UAUUUUAUCA___AGCUCCUAAUUAGGUUGC_CCAUUAAAUGGCAUAACACUUAAGAUACGCACUGUAUUGUGUUGGAUAAAAAUAAAUA .............................(((((((........(((....))).....((((...))))..((((((....(((((.....))))))))))))))))))....... (-10.36 = -10.56 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:30 2006