
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,880,087 – 11,880,278 |
| Length | 191 |
| Max. P | 0.998741 |

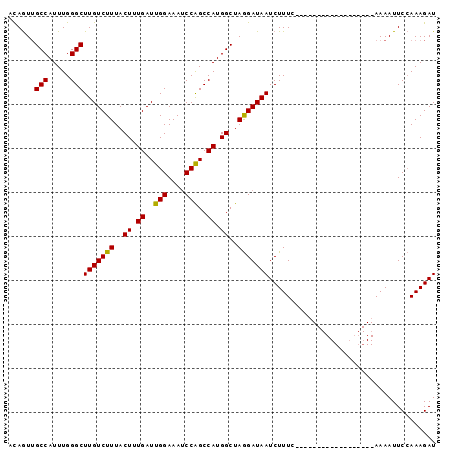
| Location | 11,880,087 – 11,880,184 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 80.95 |
| Mean single sequence MFE | -20.27 |
| Consensus MFE | -15.41 |
| Energy contribution | -14.97 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11880087 97 + 22224390 CCAAUAGCCAUUUGGGCUUGUCUUUACUUUGAUUGGAAAGCCGGCCAUGGCUAAGAUAAUCUUUCGACACGGCUUUUCACCGGAAAAUUCAAAAGAU .....((((.....)))).((((((........((((((((((((....))...((.......))....))))))))))...((....)).)))))) ( -23.20) >DroSec_CAF1 12096 79 + 1 AUAGUUGCCAUUUGGGCUUGUCUUUACUUUGAUUGGAAAUCCAGCCAUGGCUAGGAUAAUCUUUC------------------AAAAUUCCAAAGAU ......(((.....)))..((((((..(((((..((..(((((((....))).))))..))..))------------------))).....)))))) ( -18.80) >DroSim_CAF1 14433 79 + 1 ACAGUUGCCAUUUGGGCUUGUCUUUACUUUGAUUGGAAAUCCAGCCAUGGCUAGGAUAAUCUUUC------------------AAAAUUCCAAAGAU ......(((.....)))..((((((..(((((..((..(((((((....))).))))..))..))------------------))).....)))))) ( -18.80) >consensus ACAGUUGCCAUUUGGGCUUGUCUUUACUUUGAUUGGAAAUCCAGCCAUGGCUAGGAUAAUCUUUC__________________AAAAUUCCAAAGAU ......(((.....)))(((((((..((.((..(((....)))..)).))..)))))))...................................... (-15.41 = -14.97 + -0.44)


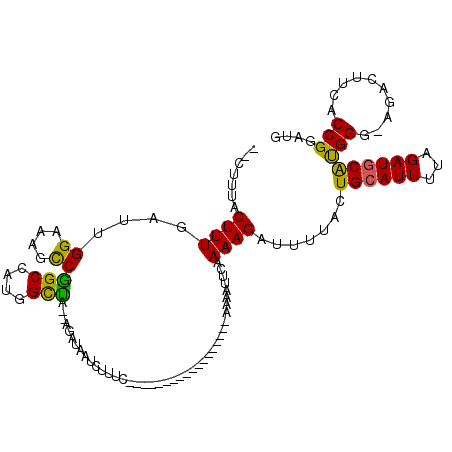
| Location | 11,880,108 – 11,880,223 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.49 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -10.42 |
| Energy contribution | -9.42 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.37 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11880108 115 + 22224390 --CUUUACUUUGAUUGGAAAGCCGGCCAUGGCUA--AGAUAAUCUUUCGACACGGCUUUUCACCGGAAAAUUCAAAAGAUUUUCCUGCAUUUUAGAUGCAUGGG-AGACUUCACCGGAUG --............((((((((((((....))..--.((.......))....))))))))))((((.....((....))((((((((((((...)))))).)))-))).....))))... ( -33.30) >DroSec_CAF1 12117 97 + 1 --CUUUACUUUGAUUGGAAAUCCAGCCAUGGCUA--GGAUAAUCUUUC------------------AAAAUUCCAAAGAUUUUACUGCAUUUUAGAUUCAUGGU-AGACUUCACCGGAUG --......(((((..((..(((((((....))).--))))..))..))------------------)))..(((...((.((((........)))).))..(((-.(....))))))).. ( -19.50) >DroSim_CAF1 14454 97 + 1 --CUUUACUUUGAUUGGAAAUCCAGCCAUGGCUA--GGAUAAUCUUUC------------------AAAAUUCCAAAGAUUUUACUGCAUUUUAGAUGCAUGGG-AGACUUCACCGGAUG --......(((((..((..(((((((....))).--))))..))..))------------------)))..(((...........((((((...)))))).((.-...)).....))).. ( -22.30) >DroEre_CAF1 14249 103 + 1 UACUCCACUUUGAUUGGUAAGUCGGCCAUGGCCA--AGAUAAUCUUAC---------------GCAAAAGUGCAAAAGAUUUUUCUGCAUUUUAGAUGCGUGGGUAGACUUCACCGGUUG ...........(((((((((((((((....))).--.......(((((---------------(((((((((((...((....))))))))))...))))))))..))))).))))))). ( -34.70) >DroYak_CAF1 15756 103 + 1 UGCUUUACUUUGAUUGGAGAGUCGGCCAUGGUCGUAUGGCUAU-UUAC---------------CGGAAAAUUCAAAAGAUUUUUCUGCAUUUUAGAUGCACGGG-AGACUUUACCGGAUG .............(((((((((((((((((....)))))))..-...(---------------(((((((.((....)).)))))((((((...))))))))).-.)))))).))))... ( -30.50) >consensus __CUUUACUUUGAUUGGAAAGCCGGCCAUGGCUA__AGAUAAUCUUUC__________________AAAAUUCAAAAGAUUUUACUGCAUUUUAGAUGCAUGGG_AGACUUCACCGGAUG .......((((....((....))(((....))).........................................)))).......((((((...))))))(((..........))).... (-10.42 = -9.42 + -1.00)

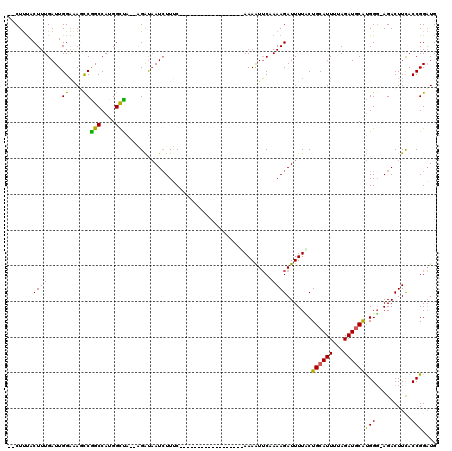
| Location | 11,880,184 – 11,880,278 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -21.63 |
| Energy contribution | -21.35 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11880184 94 + 22224390 UUUCCUGCAUUUUAGAUGCAUGGG-AGACUUCACCGGAUGGCAAUUAGCAUUUCGGUUUUGGUUUCAGUAUUUAUUUCAGCUUUGCCGCCGGUGU (((((((((((...)))))).)))-))....((((((.((((((..(((.....(((.(((....))).))).......))))))))))))))). ( -28.70) >DroSec_CAF1 12175 94 + 1 UUUACUGCAUUUUAGAUUCAUGGU-AGACUUCACCGGAUGGCAAUUAGCAUUUCGGUUUGGGUUUCAGUAUUUAUUUCAGCUUUGCCGCCGGUGU ....((((..............))-))....((((((.((((((..(((.....((..(((((......)))))..)).))))))))))))))). ( -22.94) >DroSim_CAF1 14512 94 + 1 UUUACUGCAUUUUAGAUGCAUGGG-AGACUUCACCGGAUGGCAAUUAGCAUUUCGGUUUGGGUUUCAGUAUUUAUUUCAGCUUUGCCGCCGGUGU .....((((((...)))))).((.-...)).((((((.((((((..(((.....((..(((((......)))))..)).))))))))))))))). ( -26.90) >DroEre_CAF1 14312 95 + 1 UUUUCUGCAUUUUAGAUGCGUGGGUAGACUUCACCGGUUGGUAAUUAGCAUUUCGGUUUUGGUUUCAGUAUUUAUUUCAGCUUUGCCGCCGGUGU ..(((((((((...)))))).))).......(((((((.(((((..(((.....(((.(((....))).))).......))))))))))))))). ( -21.30) >DroYak_CAF1 15820 94 + 1 UUUUCUGCAUUUUAGAUGCACGGG-AGACUUUACCGGAUGGCAAUUAGCAUUUUGGUUUUGGUUUCAGUAUUUAUUUCAGCUUUGCCGCCGGUGU ((..(((((((...)))))).)..-))....((((((.((((((..(((....(((..(..((......))..)..)))))))))))))))))). ( -25.30) >consensus UUUACUGCAUUUUAGAUGCAUGGG_AGACUUCACCGGAUGGCAAUUAGCAUUUCGGUUUUGGUUUCAGUAUUUAUUUCAGCUUUGCCGCCGGUGU .....((((((...))))))...........((((((.((((((..(((..............................))))))))))))))). (-21.63 = -21.35 + -0.28)



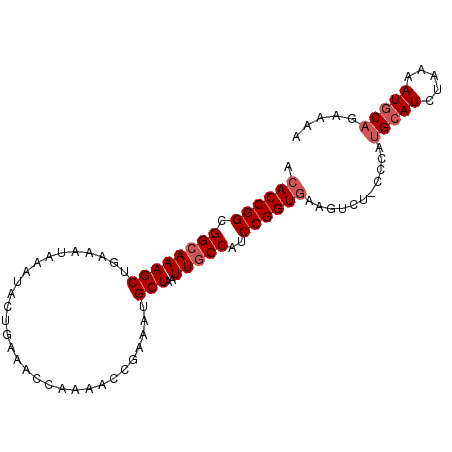
| Location | 11,880,184 – 11,880,278 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -19.95 |
| Consensus MFE | -17.79 |
| Energy contribution | -18.59 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11880184 94 - 22224390 ACACCGGCGGCAAAGCUGAAAUAAAUACUGAAACCAAAACCGAAAUGCUAAUUGCCAUCCGGUGAAGUCU-CCCAUGCAUCUAAAAUGCAGGAAA .((((((.((((((((..............................)))..)))))..))))))......-.((.(((((.....)))))))... ( -24.71) >DroSec_CAF1 12175 94 - 1 ACACCGGCGGCAAAGCUGAAAUAAAUACUGAAACCCAAACCGAAAUGCUAAUUGCCAUCCGGUGAAGUCU-ACCAUGAAUCUAAAAUGCAGUAAA .((((((.((((((((..............................)))..)))))..))))))......-........................ ( -18.41) >DroSim_CAF1 14512 94 - 1 ACACCGGCGGCAAAGCUGAAAUAAAUACUGAAACCCAAACCGAAAUGCUAAUUGCCAUCCGGUGAAGUCU-CCCAUGCAUCUAAAAUGCAGUAAA .((((((.((((((((..............................)))..)))))..))))))......-....(((((.....)))))..... ( -22.31) >DroEre_CAF1 14312 95 - 1 ACACCGGCGGCAAAGCUGAAAUAAAUACUGAAACCAAAACCGAAAUGCUAAUUACCAACCGGUGAAGUCUACCCACGCAUCUAAAAUGCAGAAAA .((((((((((...))))..........((....))......................))))))............((((.....))))...... ( -15.30) >DroYak_CAF1 15820 94 - 1 ACACCGGCGGCAAAGCUGAAAUAAAUACUGAAACCAAAACCAAAAUGCUAAUUGCCAUCCGGUAAAGUCU-CCCGUGCAUCUAAAAUGCAGAAAA ..(((((.((((((((..............................)))..)))))..))))).......-....(((((.....)))))..... ( -19.01) >consensus ACACCGGCGGCAAAGCUGAAAUAAAUACUGAAACCAAAACCGAAAUGCUAAUUGCCAUCCGGUGAAGUCU_CCCAUGCAUCUAAAAUGCAGAAAA .((((((.((((((((..............................)))..)))))..))))))...........(((((.....)))))..... (-17.79 = -18.59 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:27 2006