
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,879,790 – 11,879,938 |
| Length | 148 |
| Max. P | 0.958056 |

| Location | 11,879,790 – 11,879,898 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.91 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -30.77 |
| Energy contribution | -31.77 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
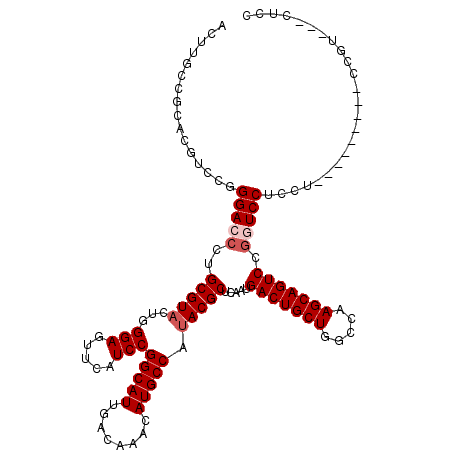
>X_DroMel_CAF1 11879790 108 + 22224390 ACUUGCCGCACGUCGGGGACCCUGCGUAUUGGGAGUUCAUCCGGCAUUGACAAACAUGCCAUACGCUCAAUGACUGCUGGCCAAGCAGUCCGUUCCUCCU---------CCGU---CUCG ...........(.((((((....((((((..(((.....)))(((((........))))))))))).....(((((((.....)))))))......))).---------))).---)... ( -35.70) >DroSec_CAF1 11808 108 + 1 ACUUGCCGCACGUCCGGGACCCUGCGUACUGGGAGUUCAUCCGGCAUUGACAAACAUGCCAUACGCUCAAUGACUGCUGGCCAAGCAGUCCGGUCCUCUU---------CCGU---CUCC .......(.(((...((((((..(((((...(((.....)))(((((........))))).))))).....(((((((.....))))))).))))))...---------.)))---)... ( -37.50) >DroSim_CAF1 14145 108 + 1 ACUUGCCGCACGUCCGGGACCCUGCGUACUGGGAGUUCAUCCGGCAUUGACAAACAUGCCAUACGCUCAAUGACUGCUGGCCAAGCAGUCCGGUCCUCCU---------CCGU---CUCC .......(.(((...((((((..(((((...(((.....)))(((((........))))).))))).....(((((((.....))))))).))))))...---------.)))---)... ( -37.50) >DroYak_CAF1 15402 120 + 1 ACUUGCCGCACGUCCGGGAGCCUGCGUUCUGGGAGUUCAUCCGGCAUUGACAAACAUGCCACACGCUCAAUGACUGCUGUCCAAGCAGUCCAGUCCACCUUCAUCUCCACCGUCUCCUCC .......(.(((....((((...((((....(((.....)))(((((........)))))..)))).....(((((((.....)))))))..............))))..))))...... ( -31.20) >consensus ACUUGCCGCACGUCCGGGACCCUGCGUACUGGGAGUUCAUCCGGCAUUGACAAACAUGCCAUACGCUCAAUGACUGCUGGCCAAGCAGUCCGGUCCUCCU_________CCGU___CUCC ................(((((..(((((...(((.....)))(((((........))))).))))).....(((((((.....))))))).)))))........................ (-30.77 = -31.77 + 1.00)

| Location | 11,879,830 – 11,879,938 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.01 |
| Mean single sequence MFE | -38.01 |
| Consensus MFE | -30.33 |
| Energy contribution | -30.89 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.958056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
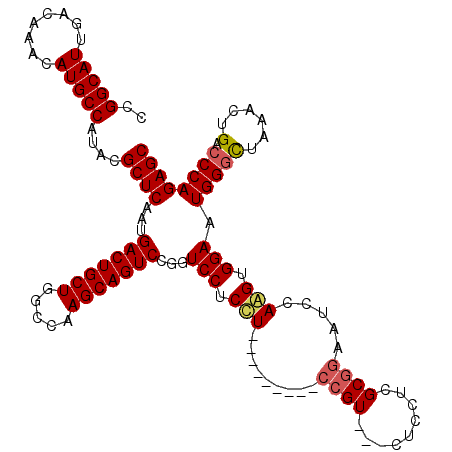
>X_DroMel_CAF1 11879830 108 + 22224390 CCGGCAUUGACAAACAUGCCAUACGCUCAAUGACUGCUGGCCAAGCAGUCCGUUCCUCCU---------CCGU---CUCGUCGCGGAAUCCAAGUGGAAUGGACUAAACUGACCCAGAGC ..(((((........)))))....((((.......(((.....)))((((((((((.(.(---------((((---......)))))......).))))))))))...........)))) ( -35.00) >DroSec_CAF1 11848 108 + 1 CCGGCAUUGACAAACAUGCCAUACGCUCAAUGACUGCUGGCCAAGCAGUCCGGUCCUCUU---------CCGU---CUCCUCGCGGAAUCCAAGUGGAAUGGGCUAAACUGACCCAGAGC ..(((((........)))))....((((...(((((((.....))))))).(((((..((---------((((---......))))))(((....)))..)))))...........)))) ( -38.20) >DroSim_CAF1 14185 108 + 1 CCGGCAUUGACAAACAUGCCAUACGCUCAAUGACUGCUGGCCAAGCAGUCCGGUCCUCCU---------CCGU---CUCCUCGCGGAAUCCAGGUGGAAUGGGCUAAACUGACCCAGAGC ..(((((........)))))....((((...(((((((.....)))))))...(((.(((---------((((---......))))).....)).))).(((((......).)))))))) ( -38.50) >DroYak_CAF1 15442 120 + 1 CCGGCAUUGACAAACAUGCCACACGCUCAAUGACUGCUGUCCAAGCAGUCCAGUCCACCUUCAUCUCCACCGUCUCCUCCUCGCAUGAACCAGGUGGAAUGGGUUAAAGUGACCCAGAGC ..(((((........)))))....((((...(((((((.....)))))))...((((((((((((.................).))))...))))))).((((((.....)))))))))) ( -40.33) >consensus CCGGCAUUGACAAACAUGCCAUACGCUCAAUGACUGCUGGCCAAGCAGUCCGGUCCUCCU_________CCGU___CUCCUCGCGGAAUCCAAGUGGAAUGGGCUAAACUGACCCAGAGC ..(((((........)))))....((((...(((((((.....)))))))...(((.(((.........((((.........)))).....))).))).(((((......).)))))))) (-30.33 = -30.89 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:23 2006