| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,876,262 – 11,876,364 |
| Length | 102 |
| Max. P | 0.660935 |

| Location | 11,876,262 – 11,876,364 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -17.13 |
| Energy contribution | -17.72 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11876262 102 + 22224390 UCAUCGGGUAGGUGGAAUUCGUUCCAC-GGGGGCUUUGCAUACAUUUU--CC-CUGACUCAGCCCGGACAACAUAUUUUAGGCUCUCCAUUUCCAUCUCCAUCCGC--- ....(((((((((((((.........(-(((((...((....))...)--))-)))....((((..((........))..))))......))))))))..))))).--- ( -28.30) >DroSec_CAF1 9444 104 + 1 UCAACGGGUAGGUGGAAUUCGUUCCA--GGGGGCUUUGCAUACAUUUU--CC-CUGACUCAGCCCGGACAACAUAUUUUAGGCUCUCCAUUUCCAUCUCCAUCCGCAGC ....(((((((((((((.......((--(((((...((....))..))--))-)))....((((..((........))..))))......))))))))..))))).... ( -29.40) >DroSim_CAF1 10590 104 + 1 UCAUCGGGUAGGUGGAAUUCGUUCCA--GGGGGCUUUGCAUACAUUUU--CC-CUGACUCAGCCCGGACAACAUAUUUUAGGCUCUCCAUUUCCAUCUCCAUCCGCAGC ....(((((((((((((.......((--(((((...((....))..))--))-)))....((((..((........))..))))......))))))))..))))).... ( -29.00) >DroEre_CAF1 11428 99 + 1 UCGUCGGCUAGGUGGAAUCCGUUCCA--GGGGGCUUUGCAUACAUUUU--CC-CUGACUCAGCCCGGACAACAUAUUUUAGGCUCUCCAUUUCCAUCUCCACCU----- .....((..((((((((.......((--(((((...((....))..))--))-)))....((((..((........))..))))......))))))))))....----- ( -25.70) >DroYak_CAF1 9734 99 + 1 UCGUUGGGUAGGUGGAAUCCGUACCA--GGGGGCUUUGCAUACAUUUU--CC-CUGACUCAGCCCGGACAACAUAUUUUAGGCUCUCCAUUUCUAUCUCCAUCU----- ......((.((((((((.......((--(((((...((....))..))--))-)))....((((..((........))..))))......))))))))))....----- ( -25.00) >DroMoj_CAF1 27091 87 + 1 UCUG-----UGCUGUAGCCAGCCCCAUUGGGGACUUUGCAUACAUUUUGCCCACUGGCCUGGCUUGGGCGA-AUAUUUUAGGCUCUCGCACUU---------------- ...(-----(((...((((((((((...)))).))..........((((((((..(((...))))))))))-).......))))...))))..---------------- ( -30.10) >consensus UCAUCGGGUAGGUGGAAUCCGUUCCA__GGGGGCUUUGCAUACAUUUU__CC_CUGACUCAGCCCGGACAACAUAUUUUAGGCUCUCCAUUUCCAUCUCCAUCC_____ .....((((((((((((...(((((....)))))..........................((((..((........))..))))......))))))))..))))..... (-17.13 = -17.72 + 0.59)


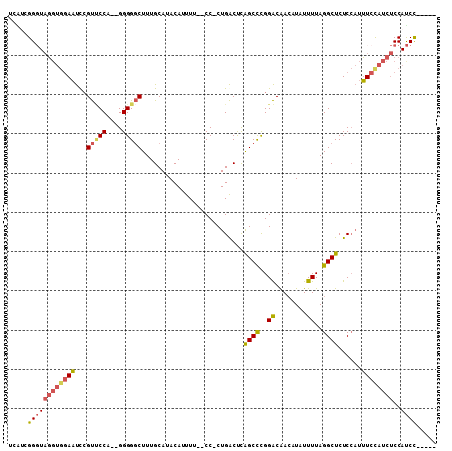
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:21 2006