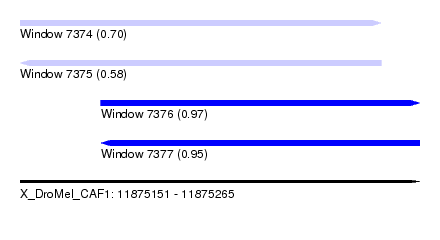
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,875,151 – 11,875,265 |
| Length | 114 |
| Max. P | 0.970575 |

| Location | 11,875,151 – 11,875,254 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.34 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -27.79 |
| Energy contribution | -28.35 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
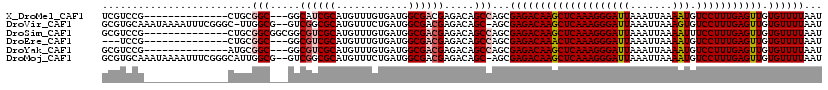
>X_DroMel_CAF1 11875151 103 + 22224390 UCGUCCG--------------CUGCGGC---GGCAUCGCAUGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAGCUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAU ((((..(--------------((((..(---(.(((((((....))))))).))....).))))..))))((((((((((((((((((.......))).))))))))))).))))..... ( -38.90) >DroVir_CAF1 23237 116 + 1 GCGUGCAAAUAAAAUUUCGGGC-UUGGCG--GUCGGCGCAUGUUUCUGAUGGCGACGAGACAGC-AGCGAGACAAGCUCAAAGGGAUUAAAUUAAAGUGUCCUUUGAGUUGUGUUUUAAU ((.(((............(.((-(.....--...))).).((((((..........))))))))-)))((((((((((((((((((((.......))).))))))))))).))))))... ( -31.60) >DroSim_CAF1 8386 106 + 1 GCGUCCG--------------CUGCGGCGGCGGCGUCGCAUGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAGCUCAAAGGGAUUAAAUUAAAAUUUCCUUUGAGUUGUGUUUUAAU ((....(--------------((((..((.((.(((((((....))))))).)).)).).))))..))((((((((((((((((((............)))))))))))).))))))... ( -42.40) >DroEre_CAF1 10331 100 + 1 ---UCCG--------------CUGCGGC---GGCGUCGCAUGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAACUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAU ---...(--------------((((..(---(.(((((((....))))))).))....).))))....((((((((((((((((((((.......))).))))))))))).))))))... ( -36.30) >DroYak_CAF1 8558 103 + 1 GCGUCCG--------------AUGCGGC---GGCGUCGCAUGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAGCUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAU (((((..--------------...((.(---(.(((((((....))))))).)).)).))).))....((((((((((((((((((((.......))).))))))))))).))))))... ( -35.30) >DroMoj_CAF1 24788 117 + 1 GCGUGCAAAUAAAAUUUCGGGCAUUGGCG--GUCGGCGCAUGUUUCUGAUGGCGACGAGACAGC-AGCGAGACAAGCUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAU ((.(((..............((....)).--((((.((((......)).)).))))......))-)))((((((((((((((((((((.......))).))))))))))).))))))... ( -32.60) >consensus GCGUCCG______________CUGCGGC___GGCGUCGCAUGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAGCUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAU .........................(((.....((((((............)))))).....)))...((((((((((((((((((((.......))).))))))))))).))))))... (-27.79 = -28.35 + 0.56)

| Location | 11,875,151 – 11,875,254 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.34 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -20.78 |
| Energy contribution | -21.09 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11875151 103 - 22224390 AUUAAAACACAACUCAAAGGACAUUUUAAUUUAAUCCCUUUGAGCUUGUCUCGCUGGCUGUCUCGUCGCCAUCACAAACAUGCGAUGCC---GCCGCAG--------------CGGACGA ............((((((((................)))))))).(((((.(((((.(.((..((((((............))))))..---)).))))--------------))))))) ( -30.49) >DroVir_CAF1 23237 116 - 1 AUUAAAACACAACUCAAAGGACACUUUAAUUUAAUCCCUUUGAGCUUGUCUCGCU-GCUGUCUCGUCGCCAUCAGAAACAUGCGCCGAC--CGCCAA-GCCCGAAAUUUUAUUUGCACGC ........((((((((((((................)))))))).)))).(((..-(((.....((((((((.......))).).))))--.....)-)).)))................ ( -21.69) >DroSim_CAF1 8386 106 - 1 AUUAAAACACAACUCAAAGGAAAUUUUAAUUUAAUCCCUUUGAGCUUGUCUCGCUGGCUGUCUCGUCGCCAUCACAAACAUGCGACGCCGCCGCCGCAG--------------CGGACGC ............((((((((................))))))))...(((.(((((((.((..((((((............))))))..)).)))..))--------------))))).. ( -33.39) >DroEre_CAF1 10331 100 - 1 AUUAAAACACAACUCAAAGGACAUUUUAAUUUAAUCCCUUUGAGUUUGUCUCGCUGGCUGUCUCGUCGCCAUCACAAACAUGCGACGCC---GCCGCAG--------------CGGA--- ......(((.((((((((((................))))))))))))).((((((.(.((..((((((............))))))..---)).))))--------------))).--- ( -31.29) >DroYak_CAF1 8558 103 - 1 AUUAAAACACAACUCAAAGGACAUUUUAAUUUAAUCCCUUUGAGCUUGUCUCGCUGGCUGUCUCGUCGCCAUCACAAACAUGCGACGCC---GCCGCAU--------------CGGACGC ............((((((((................))))))))...(((.((...((.((..((((((............))))))..---)).))..--------------))))).. ( -26.69) >DroMoj_CAF1 24788 117 - 1 AUUAAAACACAACUCAAAGGACAUUUUAAUUUAAUCCCUUUGAGCUUGUCUCGCU-GCUGUCUCGUCGCCAUCAGAAACAUGCGCCGAC--CGCCAAUGCCCGAAAUUUUAUUUGCACGC ........((((((((((((................)))))))).)))).(((.(-(((((.((..........)).))).))).))).--.((.((((..........)))).)).... ( -20.39) >consensus AUUAAAACACAACUCAAAGGACAUUUUAAUUUAAUCCCUUUGAGCUUGUCUCGCUGGCUGUCUCGUCGCCAUCACAAACAUGCGACGCC___GCCGCAG______________CGGACGC ........((((((((((((................)))))))).)))).....((((.....((((((............)))))).....))))........................ (-20.78 = -21.09 + 0.31)


| Location | 11,875,174 – 11,875,265 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 93.38 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -22.88 |
| Energy contribution | -22.93 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11875174 91 + 22224390 UGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAGCUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUC-------------- .....((((((((.........)))...((((((((((((((((((((.......))).))))))))))).)))))).)))))........-------------- ( -24.50) >DroVir_CAF1 23274 104 + 1 UGUUUCUGAUGGCGACGAGACAGC-AGCGAGACAAGCUCAAAGGGAUUAAAUUAAAGUGUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUCUCUGGCUCAAGUCA .........((((...(((.(((.-((.(((...((((((((((((((.......))).)))))))))))(((.......)))...))).))))).)))..)))) ( -27.60) >DroSec_CAF1 8432 91 + 1 UGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAGCUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUC-------------- .....((((((((.........)))...((((((((((((((((((((.......))).))))))))))).)))))).)))))........-------------- ( -24.50) >DroSim_CAF1 8412 91 + 1 UGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAGCUCAAAGGGAUUAAAUUAAAAUUUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUC-------------- .....((((((((.........)))...((((((((((((((((((............)))))))))))).)))))).)))))........-------------- ( -26.10) >DroEre_CAF1 10351 91 + 1 UGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAACUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUC-------------- .....((((((((.........)))...((((((((((((((((((((.......))).))))))))))).)))))).)))))........-------------- ( -24.50) >DroYak_CAF1 8581 91 + 1 UGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAGCUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUC-------------- .....((((((((.........)))...((((((((((((((((((((.......))).))))))))))).)))))).)))))........-------------- ( -24.50) >consensus UGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAGCUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUC______________ .....((((((((.........)))...((((((((((((((((((((.......))).))))))))))).)))))).)))))...................... (-22.88 = -22.93 + 0.06)


| Location | 11,875,174 – 11,875,265 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 93.38 |
| Mean single sequence MFE | -20.22 |
| Consensus MFE | -18.95 |
| Energy contribution | -18.95 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11875174 91 - 22224390 --------------GAGAGGAAGUAAUUAAAACACAACUCAAAGGACAUUUUAAUUUAAUCCCUUUGAGCUUGUCUCGCUGGCUGUCUCGUCGCCAUCACAAACA --------------(((((..(((.........((((((((((((................)))))))).)))).......)))..))).))............. ( -18.98) >DroVir_CAF1 23274 104 - 1 UGACUUGAGCCAGAGAGAGGAAGUAAUUAAAACACAACUCAAAGGACACUUUAAUUUAAUCCCUUUGAGCUUGUCUCGCU-GCUGUCUCGUCGCCAUCAGAAACA .(((..(((.(((((.(((...((.......))((((((((((((................)))))))).))))))).))-.))).))))))............. ( -24.39) >DroSec_CAF1 8432 91 - 1 --------------GAGAGGAAGUAAUUAAAACACAACUCAAAGGACAUUUUAAUUUAAUCCCUUUGAGCUUGUCUCGCUGGCUGUCUCGUCGCCAUCACAAACA --------------(((((..(((.........((((((((((((................)))))))).)))).......)))..))).))............. ( -18.98) >DroSim_CAF1 8412 91 - 1 --------------GAGAGGAAGUAAUUAAAACACAACUCAAAGGAAAUUUUAAUUUAAUCCCUUUGAGCUUGUCUCGCUGGCUGUCUCGUCGCCAUCACAAACA --------------(((((..(((.........((((((((((((................)))))))).)))).......)))..))).))............. ( -18.98) >DroEre_CAF1 10351 91 - 1 --------------GAGAGGAAGUAAUUAAAACACAACUCAAAGGACAUUUUAAUUUAAUCCCUUUGAGUUUGUCUCGCUGGCUGUCUCGUCGCCAUCACAAACA --------------(((((..(((.......(((.((((((((((................))))))))))))).......)))..))).))............. ( -21.03) >DroYak_CAF1 8581 91 - 1 --------------GAGAGGAAGUAAUUAAAACACAACUCAAAGGACAUUUUAAUUUAAUCCCUUUGAGCUUGUCUCGCUGGCUGUCUCGUCGCCAUCACAAACA --------------(((((..(((.........((((((((((((................)))))))).)))).......)))..))).))............. ( -18.98) >consensus ______________GAGAGGAAGUAAUUAAAACACAACUCAAAGGACAUUUUAAUUUAAUCCCUUUGAGCUUGUCUCGCUGGCUGUCUCGUCGCCAUCACAAACA ..............(((((..(((.........((((((((((((................)))))))).)))).......)))..))).))............. (-18.95 = -18.95 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:19 2006