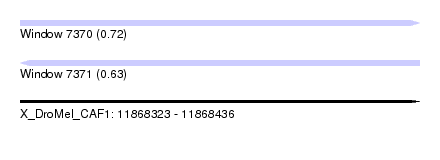
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,868,323 – 11,868,436 |
| Length | 113 |
| Max. P | 0.722337 |

| Location | 11,868,323 – 11,868,436 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.18 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -27.08 |
| Energy contribution | -27.58 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11868323 113 + 22224390 UUUUAUAUUUUACUGCAAUUU----CCAGUCCUUUGAGACGGUAUCCAAUAGCAUAAUGCUGGCCCUGCUCUGCUUGGCCACCAACAAGGGUGAUUGCCAGCGACGUUUGCUGGCGG ...........((((......----.))))((((((....(((.......(((.....)))((((..((...))..)))))))..))))))....(((((((((...))))))))). ( -32.80) >DroSec_CAF1 1704 105 + 1 U--------UUACCCCCGUUU----CCAGUCCUUUGAGACGGUGUCCAAUAGCAUAAUGCUGGCCCUGCUCUGCUUGGCCACCAACAAGGGCGACUGCCAGCGACGUUUGCUGGCGG .--------......((((((----(.........)))))))(((((...(((.....)))((((..((...))..))))........))))).((((((((((...)))))))))) ( -39.80) >DroSim_CAF1 1650 105 + 1 U--------UUACUCCCGUUU----CCAGUCCUUUGAGACGGUGUCCAAUAGCAUAAUGCUGGCCCUGCUCUGCUUGGCCACCAACAAGGGCGACUGCCAGCGACGCUUGCUGGCGG .--------......((((((----(.........)))))))(((((...(((.....)))((((..((...))..))))........))))).((((((((((...)))))))))) ( -39.80) >DroEre_CAF1 3365 100 + 1 -----------------AUUUCCUGCCAGUCCUUUGAGACGGUGUCCAAUAGCAUAAUGCUGGCCCUGCUCUGCCUGGCCACCAACAAGAGCGACUGCCAGCAACGAUUGCUGGCGG -----------------.....((((((((...(((.....((((......))))..(((((((..((((((...(((...)))...))))))...))))))).)))..)))))))) ( -38.10) >DroYak_CAF1 1609 96 + 1 -----------------AUUU----GCAGUCCUUCGAGACGGUGUCCAAUAGCAUAAUGCUGGCCCUCCUCUGCUUGGCCACCAACAAGGGCGUCUGCCAGCGUCGCUUGCUGGCGG -----------------....----...((((((...(..((((.((((((((.....))))((........)))))).))))..)))))))..(((((((((.....))))))))) ( -34.90) >DroMoj_CAF1 12890 97 + 1 ----------------UCCUU----GCAGUCCUUUGAGACCGUCUCGAAUAGCAUUAUGAUCGCGCUGCUCUGCCUGGCCACGCACAAGGGCGACUGCCAGCAGCGCCUGCGCGCGG ----------------....(----((.((..((((((.....))))))..)).........((((.((.((((.((((..(((......)))...)))))))).))..))))))). ( -37.20) >consensus _________________AUUU____CCAGUCCUUUGAGACGGUGUCCAAUAGCAUAAUGCUGGCCCUGCUCUGCUUGGCCACCAACAAGGGCGACUGCCAGCGACGCUUGCUGGCGG ............................((((((...(..((((.(((.((((.....))))((........)).))).))))..)))))))..((((((((((...)))))))))) (-27.08 = -27.58 + 0.50)

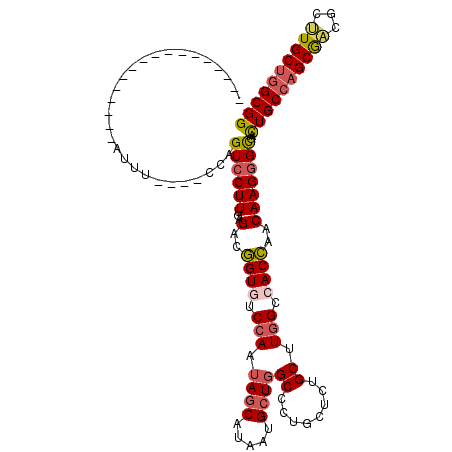
| Location | 11,868,323 – 11,868,436 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.18 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -26.53 |
| Energy contribution | -26.65 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11868323 113 - 22224390 CCGCCAGCAAACGUCGCUGGCAAUCACCCUUGUUGGUGGCCAAGCAGAGCAGGGCCAGCAUUAUGCUAUUGGAUACCGUCUCAAAGGACUGG----AAAUUGCAGUAAAAUAUAAAA ..((((((.......)))))).....((.(((.((((((((..((...))..))))(((.....))).......))))...))).))((((.----......))))........... ( -33.20) >DroSec_CAF1 1704 105 - 1 CCGCCAGCAAACGUCGCUGGCAGUCGCCCUUGUUGGUGGCCAAGCAGAGCAGGGCCAGCAUUAUGCUAUUGGACACCGUCUCAAAGGACUGG----AAACGGGGGUAA--------A ..((((((.......))))))....(((((((((((((.((((....((((..(....)....)))).)))).))))((((....))))...----.)))))))))..--------. ( -42.90) >DroSim_CAF1 1650 105 - 1 CCGCCAGCAAGCGUCGCUGGCAGUCGCCCUUGUUGGUGGCCAAGCAGAGCAGGGCCAGCAUUAUGCUAUUGGACACCGUCUCAAAGGACUGG----AAACGGGAGUAA--------A ..((((((.......))))))(.((.((.(((.(((((.((((....((((..(....)....)))).)))).)))))...))).)).(((.----...))))).)..--------. ( -37.20) >DroEre_CAF1 3365 100 - 1 CCGCCAGCAAUCGUUGCUGGCAGUCGCUCUUGUUGGUGGCCAGGCAGAGCAGGGCCAGCAUUAUGCUAUUGGACACCGUCUCAAAGGACUGGCAGGAAAU----------------- ..(((((((.....)))))))......(((((((((((.((((((........)))(((.....)))..))).))))((((....)))).)))))))...----------------- ( -39.10) >DroYak_CAF1 1609 96 - 1 CCGCCAGCAAGCGACGCUGGCAGACGCCCUUGUUGGUGGCCAAGCAGAGGAGGGCCAGCAUUAUGCUAUUGGACACCGUCUCGAAGGACUGC----AAAU----------------- (((((((((((.......(((....))))))))))))))....((((.(..((.(((((.....))...)))...))..)((....))))))----....----------------- ( -33.51) >DroMoj_CAF1 12890 97 - 1 CCGCGCGCAGGCGCUGCUGGCAGUCGCCCUUGUGCGUGGCCAGGCAGAGCAGCGCGAUCAUAAUGCUAUUCGAGACGGUCUCAAAGGACUGC----AAGGA---------------- ((((((((((((((((....))).))))..)))))))))((..((((((((............))))....(((.....)))......))))----..)).---------------- ( -40.30) >consensus CCGCCAGCAAGCGUCGCUGGCAGUCGCCCUUGUUGGUGGCCAAGCAGAGCAGGGCCAGCAUUAUGCUAUUGGACACCGUCUCAAAGGACUGG____AAAU_________________ ..((((((.......)))))).....((.(((.(((((.((((....((((..(....)....)))).)))).)))))...))).)).............................. (-26.53 = -26.65 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:14 2006