| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,327,806 – 1,327,912 |
| Length | 106 |
| Max. P | 0.518332 |

| Location | 1,327,806 – 1,327,912 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.07 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -22.77 |
| Energy contribution | -22.88 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
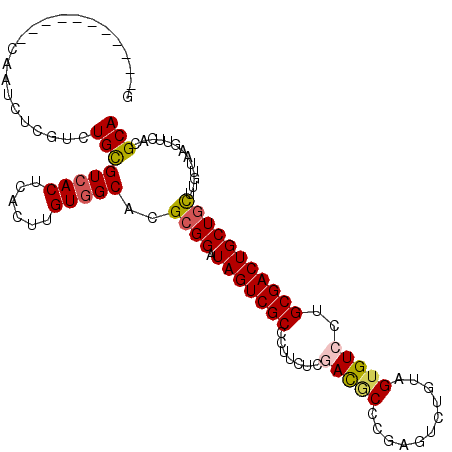
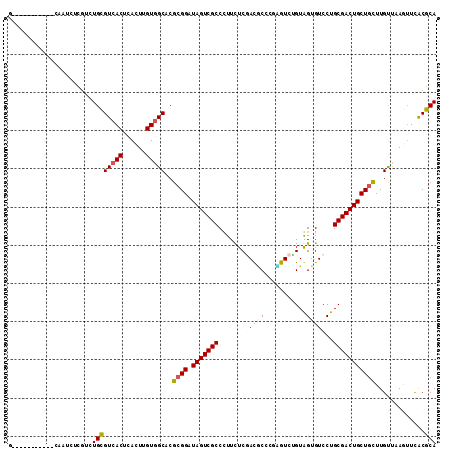
>X_DroMel_CAF1 1327806 106 - 22224390 GGCCC------GCAUGCUUGAGUGCGUCACUUACUGGUGGCCCGCGGAUAGUCGCCCUUUUCGACGCCCGAGUCCGUGGUGUCCUGCGACUGCUGCUUGUUAAGUUCGCGCA ....(------((..((((((..(.((((((....)))))).)((((.(((((((.......((((((((....)).))))))..)))))))))))...))))))..))).. ( -40.80) >DroVir_CAF1 11021 107 - 1 -----CGGUAAACAACAUUGGCUGCGUCACUCACUUGUCGCACGAGGGUAGUCGCCCUUUUCCACACCCGUGUCGGUAGUGUCCUGCGACUGCUGUUUGCCCAAAUUACGCA -----.(((((((....((.(.((((.((......)).))))).))(((((((((.......((((((......))).)))....))))))))))))))))........... ( -33.30) >DroPse_CAF1 3097 101 - 1 G-----------CCAUUGCGUCUGCGUCACUCACUUGUGGCACGCGGAUAGUCGCCUUUCUCGAUGCCCCCAUCUGUAGUGUCCUGCGACUGCUGUUUGUUUAGUUCACGCA .-----------....(((((..(((((((......)))))..((((.(((((((....(((((((....)))).).))......))))))))))).......))..))))) ( -29.90) >DroEre_CAF1 3117 106 - 1 AACCC------GCAUGCUCGAGUGCGUCACUUACUGGUGGCCCGCGGGUAGUCGCCCUUCUCGACUCCCGAGUCUGUGGUGUCCUGCGACUGCUGCUUGUUAAGUUCGCGCA .((((------(((.((((..(((.((((((....)))))).)))(((.(((((.......)))))))))))).))))).))...(((...(((........)))...))). ( -42.40) >DroWil_CAF1 534 98 - 1 --------------AUCUCAUCUGUGUCACUCACUUGUGGCUCGCGGAUAGUCGCCCUUCUCCACGCUGUUGUCAGUGGUGUCCUGCGACUGCUGCUUGUUCAAUUCACGCA --------------.........(.(((((......))))).)((((.(((((((....(.((((..........)))).)....)))))))))))................ ( -29.50) >DroPer_CAF1 2874 101 - 1 G-----------CCAUUGCGUCUGCGUCACUCACUUGUGGCACGCGGAUAGUCGCCUUUCUCGAUGCCCCCAUCUGUAGUGUCCUGCGACUGCUGUUUGUUUAGUUCACGCA .-----------....(((((..(((((((......)))))..((((.(((((((....(((((((....)))).).))......))))))))))).......))..))))) ( -29.90) >consensus G___________CAAUCUCGUCUGCGUCACUCACUUGUGGCACGCGGAUAGUCGCCCUUCUCGACGCCCGAGUCUGUAGUGUCCUGCGACUGCUGCUUGUUAAGUUCACGCA ......................((((((((......)))))..((((.(((((((.......(((((...........)))))..))))))))))).............))) (-22.77 = -22.88 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:48 2006