| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,855,092 – 11,855,189 |
| Length | 97 |
| Max. P | 0.579299 |

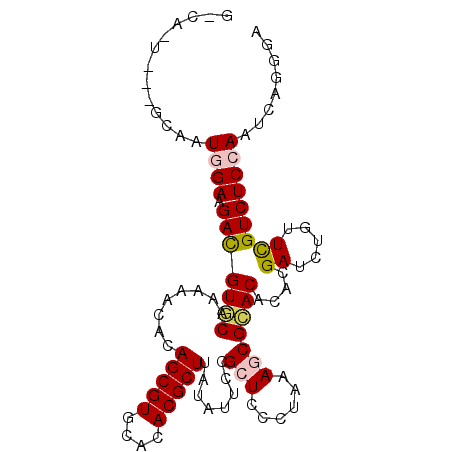
| Location | 11,855,092 – 11,855,189 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 78.34 |
| Mean single sequence MFE | -25.41 |
| Consensus MFE | -15.73 |
| Energy contribution | -15.87 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11855092 97 + 22224390 GACAAUG--GCCAUGGAAGACGUGCAGAAGCACAGCGUGCACACGCUGAUAUUCCGCUCCCUGAAGCGCACCCACGAUCUGUUCGUCUCCAACCAGGGA ..(..((--(...((((.((((.(((((....((((((....))))))......((((......)))).........))))).)))))))).)))..). ( -33.70) >DroPse_CAF1 4741 97 + 1 --UAUUUGAUCAAUGGAGGACGUGCAAAAACACAGCGUGCACACGCUUAUAUUCCGCUCCCUAAAGCGCACACACGAUCUAUUCGUCUCCAAUCAAGGA --..((((((...(((((...((((........(((((....)))))........(((......)))))))..((((.....))))))))))))))).. ( -27.60) >DroWil_CAF1 12954 91 + 1 AA--------AAAUGGAAGAUGUACAAAAGCAUAGCGUGCAUACGCUUAUCUUUCGCUCUCUUAAACGUACACAUGAUUUGUUUGUCUCAAAUCAGGGA ..--------....((((((((...........(((((....)))))))))))))..((((......(....).(((((((.......))))))))))) ( -17.10) >DroMoj_CAF1 11429 97 + 1 GGACAUA--AGCAUGGAAGAUGUACAAAAGCACAGCGUGCAUACGCUUAUAUUUAGAUCACUUAAACGCACACAUGAUCUGUUUGUCUCGAAUCAGGGU (((((((--(((.((.........))...((((...))))....))))))...(((((((..............)))))))...))))........... ( -17.34) >DroAna_CAF1 10338 94 + 1 GCCA-----GCGAUGGAGGAUGUGCAAAAACACAGCGUGCACACGCUGAUCUUCCGCUCCCUGAAGCGCACCCACGACCUGUUCGUCUCCAACCAGGGA ((((-----(.((((((((((((......)).((((((....)))))))))))))).)).)))..))...(((((((.....)))).........))). ( -29.10) >DroPer_CAF1 4812 97 + 1 --UAUUUGAUCAAUGGAGGACGUGCAAAAACACAGCGUGCACACGCUUAUAUUCCGCUCCCUAAAGCGCACACACGAUCUGUUCGUCUCCAAUCAAGGA --..((((((...(((((...((((........(((((....)))))........(((......)))))))..((((.....))))))))))))))).. ( -27.60) >consensus G_CA_U___GCAAUGGAAGACGUGCAAAAACACAGCGUGCACACGCUUAUAUUCCGCUCCCUAAAGCGCACACACGAUCUGUUCGUCUCCAAUCAGGGA .............((((.(((((((........(((((....)))))........(((......)))))))....((.....)))))))))........ (-15.73 = -15.87 + 0.14)


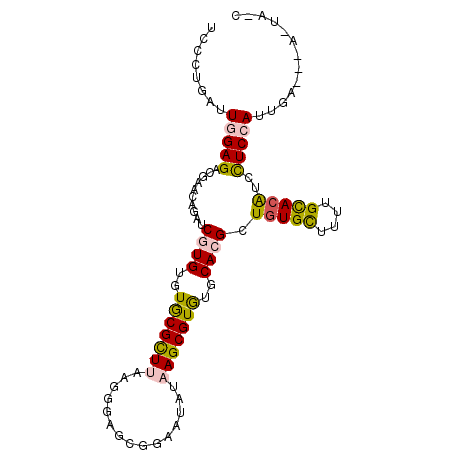
| Location | 11,855,092 – 11,855,189 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 78.34 |
| Mean single sequence MFE | -29.16 |
| Consensus MFE | -19.65 |
| Energy contribution | -19.26 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11855092 97 - 22224390 UCCCUGGUUGGAGACGAACAGAUCGUGGGUGCGCUUCAGGGAGCGGAAUAUCAGCGUGUGCACGCUGUGCUUCUGCACGUCUUCCAUGGC--CAUUGUC ....(((((((((((((.....))....((((((((....))))((((...((((((....))))))...))))))))))).))))..))--))..... ( -34.60) >DroPse_CAF1 4741 97 - 1 UCCUUGAUUGGAGACGAAUAGAUCGUGUGUGCGCUUUAGGGAGCGGAAUAUAAGCGUGUGCACGCUGUGUUUUUGCACGUCCUCCAUUGAUCAAAUA-- ...((((((((((((((.....))))..((((((((....))))((((((((.((((....)))))))))))).))))...)))))...)))))...-- ( -31.30) >DroWil_CAF1 12954 91 - 1 UCCCUGAUUUGAGACAAACAAAUCAUGUGUACGUUUAAGAGAGCGAAAGAUAAGCGUAUGCACGCUAUGCUUUUGUACAUCUUCCAUUU--------UU ....(((((((.......))))))).((((((((((....))))(((((...(((((....)))))...))))))))))).........--------.. ( -21.20) >DroMoj_CAF1 11429 97 - 1 ACCCUGAUUCGAGACAAACAGAUCAUGUGUGCGUUUAAGUGAUCUAAAUAUAAGCGUAUGCACGCUGUGCUUUUGUACAUCUUCCAUGCU--UAUGUCC .....(((..(((......((((..((((((((((((.((.......)).))))))))))))....((((....))))))))......))--)..))). ( -20.90) >DroAna_CAF1 10338 94 - 1 UCCCUGGUUGGAGACGAACAGGUCGUGGGUGCGCUUCAGGGAGCGGAAGAUCAGCGUGUGCACGCUGUGUUUUUGCACAUCCUCCAUCGC-----UGGC ...(..(((((((((((.....))))..((((((((....)))).((((((((((((....)))))).))))))))))...)))))..))-----..). ( -35.60) >DroPer_CAF1 4812 97 - 1 UCCUUGAUUGGAGACGAACAGAUCGUGUGUGCGCUUUAGGGAGCGGAAUAUAAGCGUGUGCACGCUGUGUUUUUGCACGUCCUCCAUUGAUCAAAUA-- ...(((((((((((((.((((..((((((..(((((...............)))))..))))))))))((....)).))).)))))...)))))...-- ( -31.36) >consensus UCCCUGAUUGGAGACGAACAGAUCGUGUGUGCGCUUAAGGGAGCGGAAUAUAAGCGUGUGCACGCUGUGCUUUUGCACAUCCUCCAUUGA___A_UA_C ........(((((..........((((..(((((((...............)))))))..)))).(((((....)))))..)))))............. (-19.65 = -19.26 + -0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:08 2006