
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,848,003 – 11,848,228 |
| Length | 225 |
| Max. P | 0.992413 |

| Location | 11,848,003 – 11,848,122 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 57.38 |
| Mean single sequence MFE | -20.07 |
| Consensus MFE | -13.39 |
| Energy contribution | -12.40 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.828017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11848003 119 - 22224390 UCCAAAACAGAUUGCGAAAGUGCAGCAUUACAGUUUUGAAAAGCGUAAUGAGAAGUUUUCGAGUU-UAAGCAGCUCAAAGUUUGAACACUUUUUGAACAGAAAACAGAAACUGAAGCCUA .......(((...((....))....((((((.((((....))))))))))....(((((((((((-.....)))))...(((..((.....))..))).)))))).....)))....... ( -25.10) >DroSim_CAF1 3671 118 - 1 UCCAAAACAGAUUGCGA-AGCGCAGCAUCGCAGUUUUGAAAAGCGUAAUGAGAAGUUUUCAAGUU-UAAGCACCUCAAAGNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN ..((((((....(((((-.((...)).)))))))))))..........((((..((((.......-.))))..))))........................................... ( -16.50) >DroEre_CAF1 4112 94 - 1 UUCAAGGCAGAUUGCGAAAGCGCAGCAUCACAGUUUCGAAAACACCAACGAGGCGUUUUCAAAUUAUAAGCAACUCGAGGUUUUAAG--------------------------AAGUCAA .....(((.....((....))...((.....(((((.((((((.((.....)).)))))))))))....))................--------------------------..))).. ( -18.60) >consensus UCCAAAACAGAUUGCGAAAGCGCAGCAUCACAGUUUUGAAAAGCGUAAUGAGAAGUUUUCAAGUU_UAAGCAACUCAAAGUUU_AA___________________________AAG_C_A ..((((((.(((((((....))))..)))...))))))..........((((..((((.........))))..))))........................................... (-13.39 = -12.40 + -0.99)


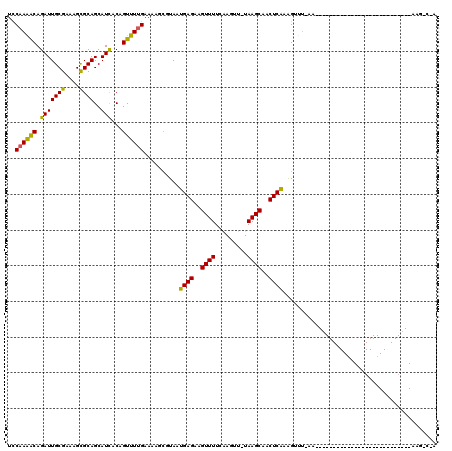
| Location | 11,848,082 – 11,848,188 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -27.45 |
| Consensus MFE | -18.55 |
| Energy contribution | -20.30 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11848082 106 + 22224390 UUCAAAACUGUAAUGCUGCACUUUCGCAAUCUGUUUUGGAAUAUUUUUUUUUUUGCAAUUUGCCAUCGGGUUAAUU--------------UGCCCGUUGGCGACUUCUGUUGCUCUGUUA ((((((((.(...(((.........)))..).))))))))..............(((((((((((.(((((.....--------------.))))).)))))).....)))))....... ( -29.30) >DroSim_CAF1 3750 100 + 1 UUCAAAACUGCGAUGCUGCGCU-UCGCAAUCUGUUUUGGAAUAUUU-----UUUGCAAUUUGCCAUCGGGUUAAUU--------------UGCCCGUUGGCGACUUCUGUUGCUCUGUUA (((((((((((((.((...)).-)))))....))))))))......-----...(((((((((((.(((((.....--------------.))))).)))))).....)))))....... ( -34.30) >DroEre_CAF1 4166 100 + 1 UUCGAAACUGUGAUGCUGCGCUUUCGCAAUCUGCCUUGAAAU--AU----UUUUGCAAUUUGCCAUCGGGUUAAUU--------------UGCCCGUUGCCGGCUACUGUAACUCUGUUA ......((.(((..((((.((....((((..(((...(((..--..----))).)))..))))...(((((.....--------------.)))))..))))))))).)).......... ( -21.90) >DroYak_CAF1 4056 114 + 1 UUCGAA--UGUGAUGCUGCACUUUCGCCAUCUGUUUUGCAAUAUAU----UUUAGCAAUUCGCCAUCGGGUUAAUUUUCCCAAUGCCCAUUGCCUGUUGCCGGCUUCUGUUGCUCUGUUA ..((((--.(((......))).))))...........((((((.(.----.(..(((((..((.((.((((.............)))))).))..)))))..)..).))))))....... ( -24.32) >consensus UUCAAAACUGUGAUGCUGCACUUUCGCAAUCUGUUUUGGAAUAUAU____UUUUGCAAUUUGCCAUCGGGUUAAUU______________UGCCCGUUGCCGACUUCUGUUGCUCUGUUA ((((((((...(((..(((......)))))).))))))))..............(((((((((((.(((((....................))))).)))))).....)))))....... (-18.55 = -20.30 + 1.75)

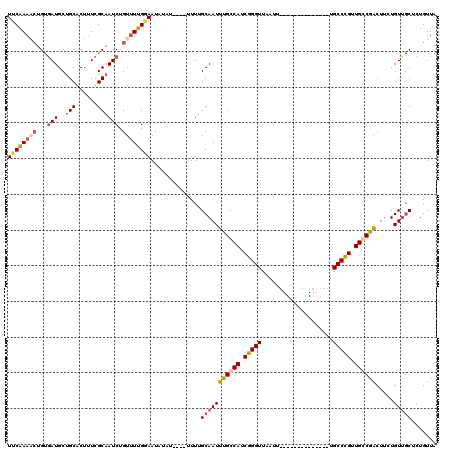
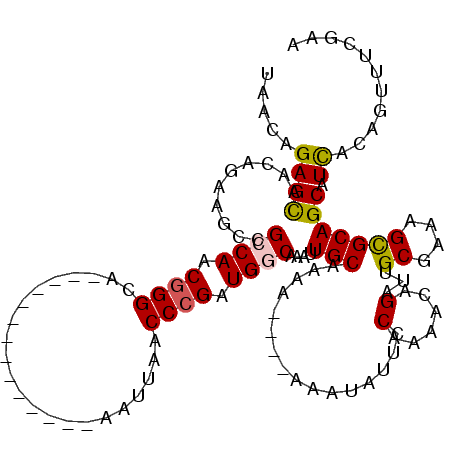
| Location | 11,848,082 – 11,848,188 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -12.95 |
| Energy contribution | -13.07 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11848082 106 - 22224390 UAACAGAGCAACAGAAGUCGCCAACGGGCA--------------AAUUAACCCGAUGGCAAAUUGCAAAAAAAAAAAUAUUCCAAAACAGAUUGCGAAAGUGCAGCAUUACAGUUUUGAA ...((((((..........((((.((((..--------------......)))).))))...((((..............((.......))..((....)))))).......)))))).. ( -24.00) >DroSim_CAF1 3750 100 - 1 UAACAGAGCAACAGAAGUCGCCAACGGGCA--------------AAUUAACCCGAUGGCAAAUUGCAAA-----AAAUAUUCCAAAACAGAUUGCGA-AGCGCAGCAUCGCAGUUUUGAA .......((((..(....)((((.((((..--------------......)))).))))...))))...-----........((((((....(((((-.((...)).))))))))))).. ( -27.80) >DroEre_CAF1 4166 100 - 1 UAACAGAGUUACAGUAGCCGGCAACGGGCA--------------AAUUAACCCGAUGGCAAAUUGCAAAA----AU--AUUUCAAGGCAGAUUGCGAAAGCGCAGCAUCACAGUUUCGAA .....(((((.(....(((..((.((((..--------------......)))).))((.....))....----..--.......))).....((....))).))).))........... ( -20.90) >DroYak_CAF1 4056 114 - 1 UAACAGAGCAACAGAAGCCGGCAACAGGCAAUGGGCAUUGGGAAAAUUAACCCGAUGGCGAAUUGCUAAA----AUAUAUUGCAAAACAGAUGGCGAAAGUGCAGCAUCACA--UUCGAA .....((((((........(....).((((((.(.(((((((........))))))).)..))))))...----.....))))......((((((....))....))))...--.))... ( -29.60) >consensus UAACAGAGCAACAGAAGCCGCCAACGGGCA______________AAUUAACCCGAUGGCAAAUUGCAAAA____AAAUAUUCCAAAACAGAUUGCGAAAGCGCAGCAUCACAGUUUCGAA .....((((..........((((.((((......................)))).))))....(((................(......)...((....))))))).))........... (-12.95 = -13.07 + 0.13)


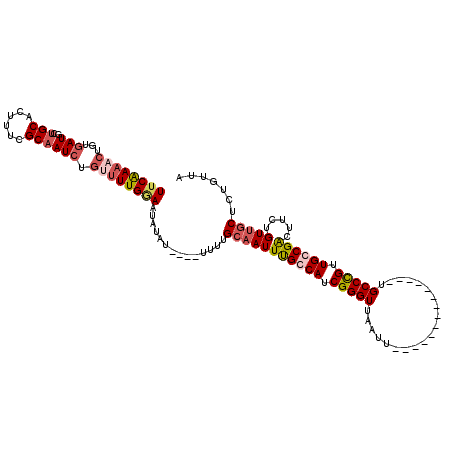
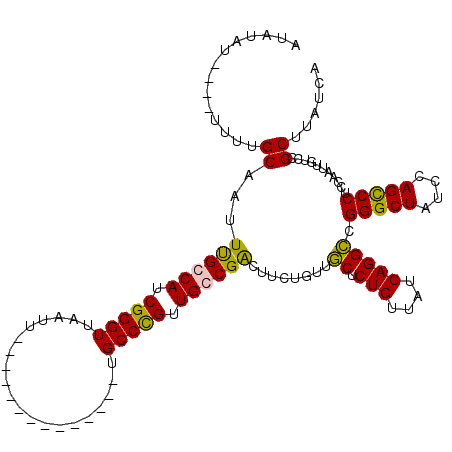
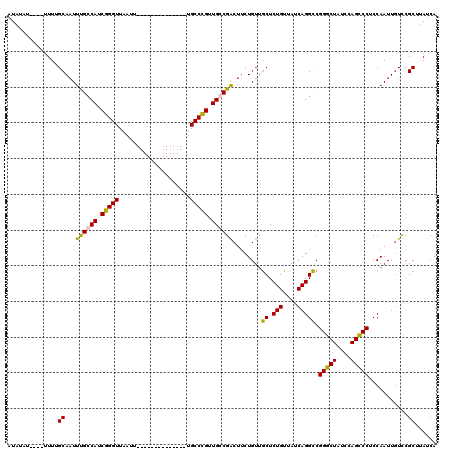
| Location | 11,848,122 – 11,848,228 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.08 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -25.19 |
| Energy contribution | -24.50 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11848122 106 + 22224390 AUAUUUUUUUUUUUGCAAUUUGCCAUCGGGUUAAUU--------------UGCCCGUUGGCGACUUCUGUUGCUCUGUUAUCAGGCCGGGCUAUCCAGCCCUCCAAUUCUCCGCUUAUCA ..............(((((((((((.(((((.....--------------.))))).)))))).....)))))((((....))))..(((((....)))))................... ( -31.40) >DroSim_CAF1 3789 101 + 1 AUAUUU-----UUUGCAAUUUGCCAUCGGGUUAAUU--------------UGCCCGUUGGCGACUUCUGUUGCUCUGUUAUCAGGCCGGGCUAUCCAGCCCUCCAAUUGUCCGCUUAUCA ......-----...((.....((((.(((((.....--------------.))))).))))(((.......((.(((....))))).(((((....))))).......))).))...... ( -32.80) >DroEre_CAF1 4206 100 + 1 AU--AU----UUUUGCAAUUUGCCAUCGGGUUAAUU--------------UGCCCGUUGCCGGCUACUGUAACUCUGUUAUCAGGUCGGGCUAUCCAGCCCUCCAAUUGUUCGCUUAUCA ..--..----....((((((.(((..(((((.....--------------.))))).....)))..(((((((...)))).)))...(((((....)))))...)))))).......... ( -25.40) >DroYak_CAF1 4094 116 + 1 AUAUAU----UUUAGCAAUUCGCCAUCGGGUUAAUUUUCCCAAUGCCCAUUGCCUGUUGCCGGCUUCUGUUGCUCUGUUAUCAGGCCGGGCUAUCCAGUCCUCCAAUUGUUCGCUUAUCA ......----...(((((((.......(((........)))...((((...(((((..((.(((.......)).).))...))))).)))).............)))))))......... ( -26.60) >consensus AUAUAU____UUUUGCAAUUUGCCAUCGGGUUAAUU______________UGCCCGUUGCCGACUUCUGUUGCUCUGUUAUCAGGCCGGGCUAUCCAGCCCUCCAAUUGUCCGCUUAUCA ..............((...((((((.(((((....................))))).))))))........((.(((....))))).(((((....)))))...........))...... (-25.19 = -24.50 + -0.69)

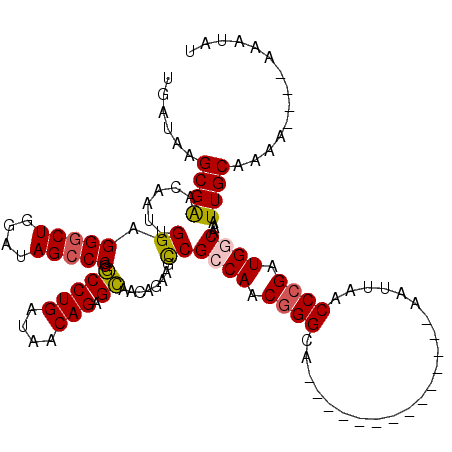
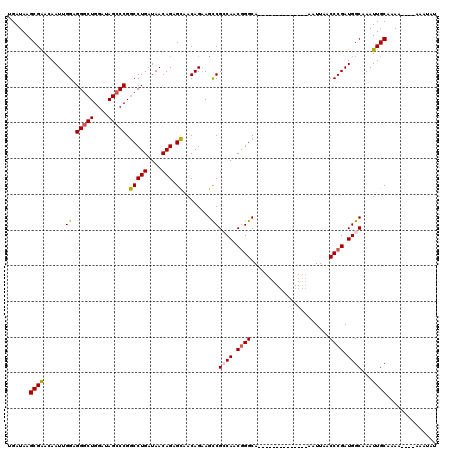
| Location | 11,848,122 – 11,848,228 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.08 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -21.92 |
| Energy contribution | -22.05 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11848122 106 - 22224390 UGAUAAGCGGAGAAUUGGAGGGCUGGAUAGCCCGGCCUGAUAACAGAGCAACAGAAGUCGCCAACGGGCA--------------AAUUAACCCGAUGGCAAAUUGCAAAAAAAAAAAUAU ......((((.((.(((..(((((....))))).(((((....))).))..)))...))((((.((((..--------------......)))).))))...)))).............. ( -31.50) >DroSim_CAF1 3789 101 - 1 UGAUAAGCGGACAAUUGGAGGGCUGGAUAGCCCGGCCUGAUAACAGAGCAACAGAAGUCGCCAACGGGCA--------------AAUUAACCCGAUGGCAAAUUGCAAA-----AAAUAU ......((((((..(((..(((((....))))).(((((....))).))..)))..)))((((.((((..--------------......)))).))))....)))...-----...... ( -33.90) >DroEre_CAF1 4206 100 - 1 UGAUAAGCGAACAAUUGGAGGGCUGGAUAGCCCGACCUGAUAACAGAGUUACAGUAGCCGGCAACGGGCA--------------AAUUAACCCGAUGGCAAAUUGCAAAA----AU--AU ......((((...((((..(((((....)))))((((((....))).))).)))).((((....((((..--------------......)))).))))...))))....----..--.. ( -29.20) >DroYak_CAF1 4094 116 - 1 UGAUAAGCGAACAAUUGGAGGACUGGAUAGCCCGGCCUGAUAACAGAGCAACAGAAGCCGGCAACAGGCAAUGGGCAUUGGGAAAAUUAACCCGAUGGCGAAUUGCUAAA----AUAUAU ..................(((.((((.....)))))))........(((((((...((((....).)))..))(.(((((((........))))))).)...)))))...----...... ( -31.50) >consensus UGAUAAGCGAACAAUUGGAGGGCUGGAUAGCCCGGCCUGAUAACAGAGCAACAGAAGCCGCCAACGGGCA______________AAUUAACCCGAUGGCAAAUUGCAAAA____AAAUAU ......((((......((.(((((....))))).(((((....))).))........))((((.((((......................)))).))))...)))).............. (-21.92 = -22.05 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:05 2006