| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,834,647 – 11,834,740 |
| Length | 93 |
| Max. P | 0.883108 |

| Location | 11,834,647 – 11,834,740 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 75.31 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -13.77 |
| Energy contribution | -14.15 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11834647 93 + 22224390 CUGCUCAGUAGAAAAAAAAUAAGCUUUUGCCAAUAUUUGGGU---AUUGUGGC--UCAUUAGAAAGGUGCCCCAUAUUGCCGAUGUGACAACAUCGGU ((((...))))...........((....))((((((..((((---(((....(--(....))...))))))).))))))(((((((....))))))). ( -23.40) >DroSec_CAF1 10515 80 + 1 CUGCUCAGUAGCAA--AAAUAAGCUU-----------UGGGU---AUUGUGGC--UUAUUAGAAAGGUGCCCCAUAUUGCCGAUGUGACAACAUCGGU .(((((((.(((..--......))))-----------)))))---).(((((.--.((((.....))))..)))))..((((((((....)))))))) ( -24.70) >DroSim_CAF1 10965 91 + 1 CUGCUCAGUAGCAA--AAAUAAGCUUUGGCCAAUAUUUGGGU---AUUGUGGC--UUAUUAGAAAGGUGCCCCAUAUUGCCGAUGUGACAACAUCGGU ..((.(((.(((..--......))))))))((((((..((((---(((....(--(....))...))))))).))))))(((((((....))))))). ( -27.70) >DroEre_CAF1 11736 81 + 1 UGGCUC-----------------CUUUUGUCGAUAUUUGGUGUGGUUGGUGCCGGCUAUUAGAAAGGUGGCCCAUAUAGCCGAUGUGACAACAUCGGU .(((((-----------------(((((...((((.((((..(.....)..)))).)))).)))))).))))......((((((((....)))))))) ( -29.80) >consensus CUGCUCAGUAGCAA__AAAUAAGCUUUUGCCAAUAUUUGGGU___AUUGUGGC__UUAUUAGAAAGGUGCCCCAUAUUGCCGAUGUGACAACAUCGGU ......................................((((....((.(((......))).))....))))......((((((((....)))))))) (-13.77 = -14.15 + 0.38)


| Location | 11,834,647 – 11,834,740 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 75.31 |
| Mean single sequence MFE | -20.30 |
| Consensus MFE | -12.14 |
| Energy contribution | -12.20 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.706398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
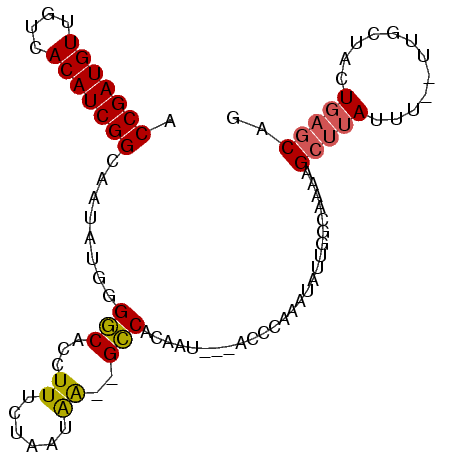
>X_DroMel_CAF1 11834647 93 - 22224390 ACCGAUGUUGUCACAUCGGCAAUAUGGGGCACCUUUCUAAUGA--GCCACAAU---ACCCAAAUAUUGGCAAAAGCUUAUUUUUUUUCUACUGAGCAG .((.((((((((.....)))))))).))((........(((((--((..((((---(......)))))......))))))).....((....)))).. ( -20.70) >DroSec_CAF1 10515 80 - 1 ACCGAUGUUGUCACAUCGGCAAUAUGGGGCACCUUUCUAAUAA--GCCACAAU---ACCCA-----------AAGCUUAUUU--UUGCUACUGAGCAG .((.((((((((.....)))))))).))..........(((((--((......---.....-----------..))))))).--.((((....)))). ( -19.32) >DroSim_CAF1 10965 91 - 1 ACCGAUGUUGUCACAUCGGCAAUAUGGGGCACCUUUCUAAUAA--GCCACAAU---ACCCAAAUAUUGGCCAAAGCUUAUUU--UUGCUACUGAGCAG .((.((((((((.....)))))))).))..........(((((--((..((((---(......)))))......))))))).--.((((....)))). ( -22.00) >DroEre_CAF1 11736 81 - 1 ACCGAUGUUGUCACAUCGGCUAUAUGGGCCACCUUUCUAAUAGCCGGCACCAACCACACCAAAUAUCGACAAAAG-----------------GAGCCA ...(((((....)))))((((((.((((.......))))))))))(((.((.......................)-----------------).))). ( -19.20) >consensus ACCGAUGUUGUCACAUCGGCAAUAUGGGGCACCUUUCUAAUAA__GCCACAAU___ACCCAAAUAUUGGCAAAAGCUUAUUU__UUGCUACUGAGCAG .(((((((....)))))))........(((...((......))..)))..........................(((((............))))).. (-12.14 = -12.20 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:55 2006