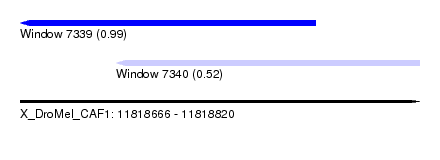
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,818,666 – 11,818,820 |
| Length | 154 |
| Max. P | 0.985196 |

| Location | 11,818,666 – 11,818,780 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 70.14 |
| Mean single sequence MFE | -40.16 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.03 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11818666 114 - 22224390 CGAUUGACCAUUAGCAACAUCAACG---GUCUGGAUGACAAUAUUGCCAUCAUCGGCGAUGGCGUCAUCCGGCGGAGCACGCACUACGGCUCGCAAGGGUCUAACAAAAGGCGCCGG .....((((...............)---)))((((((((..(((((((......)))))))..))))))))(((.....)))....((((((....))((((......)))))))). ( -41.96) >DroVir_CAF1 34763 114 - 1 CGCUUAACAAAUGCAAACGUCUUUA---GUCUGGAUGACAACAUUGCCAUCAUUGGAGAUGGCGUCAUCAGAGGCAGCACACCUCCUGGUCGGCAGCCGUUUAAACAGCGUAGACGA .((.........))...(((((...---(.(((((((((..(((..(((....)))..)))..))))))...(((.((..(((....)))..)).))).......))))..))))). ( -38.50) >DroPse_CAF1 16801 114 - 1 CGUUUGACUAACAGCCACAUCAAUA---GCCUGGAUGACAACAUAGCCAUCAUCGGAGAUGGCGUCAUCCGGAGGAGUGCACACUAUGGUUCACAGGCCUCCAAUAAACGGCGACGA ((((((.....))(((.........---....(((((((......((((((......)))))))))))))(((((.(((.((......)).)))...))))).......))))))). ( -38.50) >DroGri_CAF1 28027 114 - 1 CGCUUAACAAAUGCGAAUAUCUUUA---GUUUGGAUGACAACAUUGCCAUCAUUGGAGAUGGCGUCACCAGAUGUGGUGCACCACCCAAUCCUCAGGCGUUCAAUAAGCGCAGACGA ((((((...(((((...........---((((((.((((..(((..(((....)))..)))..))))))))))((((....))))...........)))))...))))))....... ( -34.60) >DroWil_CAF1 27621 117 - 1 CGUUUAACGAAUAGCAACAUUGUUGGAUGUCUGGAUGACAAUAUUGCCAUUAUUGGUGAUGGCGUCAUCAGGAGAAGUGCCCACUAUGGUUCCCAGGCCUCAAAUAAGCGGAGACGA .(((((.....(((((....)))))((.(((((((((((..(((..(((....)))..)))..)))))).((((..(((.....)))..))))))))).))...)))))(....).. ( -36.30) >DroMoj_CAF1 30706 114 - 1 CGCUUGGCAAGCGCAAAUACGUUCA---GUCUGGAUGACAACAUUGCCAUCAUUGGCGAUGGCGUCACCAGGGGCGGCACUCCGCCGGGCUCGCACCCUUUUGGCAAGCGUCGACGA ((((((((.(((.(...........---.(((((.((((..((((((((....))))))))..)))))))))(((((....)))))).))).))..((....))))))))....... ( -51.10) >consensus CGCUUAACAAAUACCAACAUCUUUA___GUCUGGAUGACAACAUUGCCAUCAUUGGAGAUGGCGUCAUCAGGAGCAGCACACACCAUGGUUCGCAGGCGUCUAAUAAGCGGAGACGA ((((((.......................(((((.((((......((((((......)))))))))))))))....((..((......))..))..........))))))....... (-19.00 = -19.03 + 0.03)
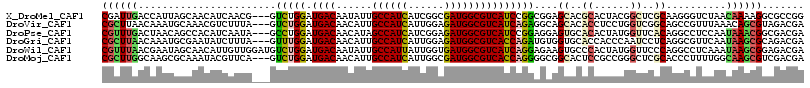
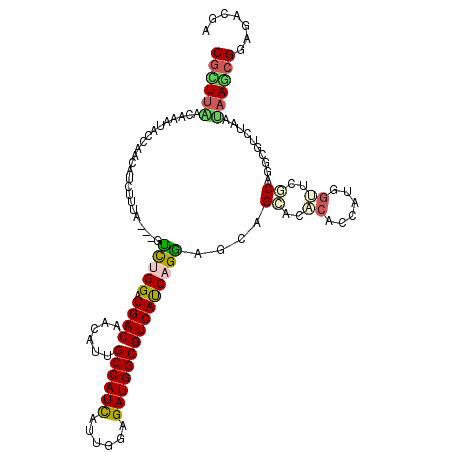

| Location | 11,818,703 – 11,818,820 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.04 |
| Mean single sequence MFE | -43.27 |
| Consensus MFE | -22.88 |
| Energy contribution | -21.63 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.53 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11818703 117 - 22224390 AGUGGGAGCUCGUCGCCGAAGGGGCUCAUCUCCUAUGGCGCGAUUGACCAUUAGCAACAUCAACG---GUCUGGAUGACAAUAUUGCCAUCAUCGGCGAUGGCGUCAUCCGGCGGAGCAC (((((..(.(((.(((((.(((((.....))))).)))))))).)..)))))...........(.---(.(((((((((..(((((((......)))))))..)))))))))).)..... ( -50.00) >DroVir_CAF1 34800 117 - 1 AGUGGGAGCUCGACGCGGAAGGGUCUCAUCACCGAUGGGACGCUUAACAAAUGCAAACGUCUUUA---GUCUGGAUGACAACAUUGCCAUCAUUGGAGAUGGCGUCAUCAGAGGCAGCAC .......(((.(((((....).((((((((...))))))))((.........))...))))....---((((.((((((..(((..(((....)))..)))..))))))..))))))).. ( -39.90) >DroPse_CAF1 16838 117 - 1 UGUGGGAGCUCGUCGCAGAAGGGGCUCAUCACCGAUGGCACGUUUGACUAACAGCCACAUCAAUA---GCCUGGAUGACAACAUAGCCAUCAUCGGAGAUGGCGUCAUCCGGAGGAGUGC .((((((((((.(......).)))))).)))).((((((..(((.....))).))))..))....---..(((((((((......((((((......)))))))))))))))........ ( -40.10) >DroGri_CAF1 28064 117 - 1 AGUGGGAGCUCGACGCAGAAGAGGCUCAUCACCGAUGGCACGCUUAACAAAUGCGAAUAUCUUUA---GUUUGGAUGACAACAUUGCCAUCAUUGGAGAUGGCGUCACCAGAUGUGGUGC .(((((..(((.........))).)))))((((((((...(((.........)))..))))....---((((((.((((..(((..(((....)))..)))..))))))))))..)))). ( -35.80) >DroWil_CAF1 27658 120 - 1 AGUGGGAGCUCGUCGCAGAAGGGGCUCAUCACCGAUGGCGCGUUUAACGAAUAGCAACAUUGUUGGAUGUCUGGAUGACAAUAUUGCCAUUAUUGGUGAUGGCGUCAUCAGGAGAAGUGC .((((((((((.(......).)))))).)))).....(((((((((((((.........))))))))).(((.((((((..(((..(((....)))..)))..)))))).)))...)))) ( -41.30) >DroMoj_CAF1 30743 117 - 1 AGUGGGUGCCCGACGCGGUAGGGUCUCAUCCCCGAUGGGACGCUUGGCAAGCGCAAAUACGUUCA---GUCUGGAUGACAACAUUGCCAUCAUUGGCGAUGGCGUCACCAGGGGCGGCAC ....(.((((((((((.(.((.((((((((...)))))))).))).)).((((......))))..---)))(((.((((..((((((((....))))))))..))))))).))))).).. ( -52.50) >consensus AGUGGGAGCUCGACGCAGAAGGGGCUCAUCACCGAUGGCACGCUUAACAAAUACCAACAUCUUUA___GUCUGGAUGACAACAUUGCCAUCAUUGGAGAUGGCGUCAUCAGGAGCAGCAC .......((((.......(((.(((.((((...)))))).).)))........................(((((.((((......((((((......))))))))))))))).).))).. (-22.88 = -21.63 + -1.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:47 2006