
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,813,824 – 11,814,024 |
| Length | 200 |
| Max. P | 0.744177 |

| Location | 11,813,824 – 11,813,944 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -26.98 |
| Energy contribution | -27.20 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.689059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11813824 120 + 22224390 AGUGCCAACACAACACAGUUGGCUAUGAUUGUCAAUAACACAGCGUAUUCAAAGGGCGGCCAUUCUAUGAUGAAUCGUGUGUACUUUCUAAUUGGGUUAUCCUCGGUUAGAAUAAACAAU ...((((((........))))))....(((((.....(((((.((.(((((.((((......))))....))))))))))))...(((((((((((.....)))))))))))...))))) ( -29.80) >DroGri_CAF1 22142 120 + 1 AGUGCCAACACAACACAGUUGGCUAUGAUUGUCAAUAACACAGCGUACUCAAAGGGCGGCCAUUCAAUUAUGAAUCGUGUGUACUUUCUAAUUGGGUUAUCCUCGGUGAGGAUAAACAAC ...((((((........))))))..((.(((((....((.....)).((((.((((..(((...((((((.(((...........))))))))))))..))))...))))))))).)).. ( -29.60) >DroWil_CAF1 21346 120 + 1 AAUGCCAACACAACACAGUUGGCAAUGAUUGUCAAUAACACAGCAUAUUCAAAGGGCGGCCAUUCUAUGAUGAAUCGUGUGUACUUUCUAAUUGGGUUAUCCUCGGUUAGGAUAAACAAU ..(((((((........))))))).((.(((((....((((.((...........)).((.((((......)))).)))))).....(((((((((.....)))))))))))))).)).. ( -30.10) >DroMoj_CAF1 23514 120 + 1 AAUGCCAACACAACACAGUUGGCUAUGAUUGUCAAUAACACAGCGUACUCAAAGGGCGGCCAUUCAAUGAUGAAUCGUGUGUACUUUCUAAUUGGGUUAUCCUCGGUUAGGAUAAACAAU ...((((((........))))))..((.(((((....((((..((..(.....)..))((.(((((....))))).)))))).....(((((((((.....)))))))))))))).)).. ( -30.60) >DroAna_CAF1 17067 120 + 1 AGCGCCAACACAACACAGUUGGCUAUGAUUGUCAAUAACACAGCGUAUUCAAAGGGCGGCCAUUCUAUGAUGAAUCGUGUGUACUUCCUAAUUGGGUUAUCCUCGGUAAGGAUGAAUAAC .((((((((........))))))..((.....))........)).((((((.((..((((.((((......)))).)).))..))((((.((((((.....)))))).)))))))))).. ( -28.50) >DroPer_CAF1 16743 120 + 1 AGCGCCAACACAACACAGUUGGCUAUGAUUGUCAAUAACACAGCGUAUUCAAAGGGCGGCCAUUCAAUGAUGAAUCGUGUGUACUUUCUAAUUGGGUUAUCCUCGGUUAGGAUAAACAAU ...((((((........))))))..((.(((((....(((((.((.(((((..((....))..((...)))))))))))))).....(((((((((.....)))))))))))))).)).. ( -30.70) >consensus AGUGCCAACACAACACAGUUGGCUAUGAUUGUCAAUAACACAGCGUAUUCAAAGGGCGGCCAUUCAAUGAUGAAUCGUGUGUACUUUCUAAUUGGGUUAUCCUCGGUUAGGAUAAACAAU ...((((((........))))))..((.(((((....((((.((.(.......).)).((.((((......)))).)))))).....(((((((((.....)))))))))))))).)).. (-26.98 = -27.20 + 0.22)



| Location | 11,813,904 – 11,814,024 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -31.82 |
| Energy contribution | -31.27 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11813904 120 + 22224390 GUACUUUCUAAUUGGGUUAUCCUCGGUUAGAAUAAACAAUGACGUCGGACCACCACCUGGUGGGUUUUCCUCUUUUUUCGGGCCACCCAUUCUAUCAGCUAAGCGAUGGGCUCCCUUUCG ((...(((((((((((.....)))))))))))...)).........((((((((....)))))....))).........(((...((((((((........)).))))))..)))..... ( -32.90) >DroGri_CAF1 22222 120 + 1 GUACUUUCUAAUUGGGUUAUCCUCGGUGAGGAUAAACAACGAAGUCGGACCACCACCUGGUGGGUUUUCUUCUUUUUUCGGGCCACCCAUUCUAUCAGCUAAGCGAUGGGCUCCCUUUCG ...........(((..((((((((...)))))))).))).((((.....(((((....))))).....)))).......(((...((((((((........)).))))))..)))..... ( -34.80) >DroEre_CAF1 17461 120 + 1 GUACUUUCUAAUUGGGUUAUCCUCGGUUAGAAUAAACAAUGACGUCGGACCACCACCUGGUGGGUUUUCCUCUUUUUUCGGGCCACCCAUUCUAUCAGCUAAGCGAUGGGCUCCCUUUCG ((...(((((((((((.....)))))))))))...)).........((((((((....)))))....))).........(((...((((((((........)).))))))..)))..... ( -32.90) >DroWil_CAF1 21426 120 + 1 GUACUUUCUAAUUGGGUUAUCCUCGGUUAGGAUAAACAAUGACGUCGGACCACCACCUGGUGGGUUAUCCUCUUUUUUUGGGCCACCCAUUCUAUCAGCUAAGCGAUGGGCUCCCUUUCG ((...(((((((((((.....)))))))))))...)).........((((((((....)))))....))).........(((...((((((((........)).))))))..)))..... ( -32.10) >DroYak_CAF1 14710 120 + 1 GUACUUUCUAAUUGGGUUAUCCUCGGUUAGAAUAAACAAUGACGUCGGACCACCACCUGGUGGGUUUUCCUCUUUUUUCGGGCCACCCAUUCUAUCAGCUAAGCGAUGGGCUCCCUUUCG ((...(((((((((((.....)))))))))))...)).........((((((((....)))))....))).........(((...((((((((........)).))))))..)))..... ( -32.90) >DroAna_CAF1 17147 120 + 1 GUACUUCCUAAUUGGGUUAUCCUCGGUAAGGAUGAAUAACGACGUCGGACCACCACCUGGUGGGUUCUCCUCUUUUUUUGGGCCACCCAUUCUAUCAGCUAAGCGAUGGGCUCCCUUUCG .....((((.((((((.....)))))).)))).((((..((....))..(((((....)))))))))............(((...((((((((........)).))))))..)))..... ( -31.50) >consensus GUACUUUCUAAUUGGGUUAUCCUCGGUUAGAAUAAACAAUGACGUCGGACCACCACCUGGUGGGUUUUCCUCUUUUUUCGGGCCACCCAUUCUAUCAGCUAAGCGAUGGGCUCCCUUUCG ((...(((((((((((.....)))))))))))...))..(((....((((((((....)))))....))).........(((...((((((((........)).))))))..)))..))) (-31.82 = -31.27 + -0.55)

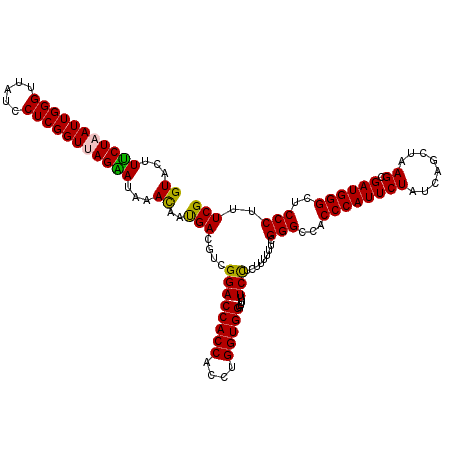

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:45 2006