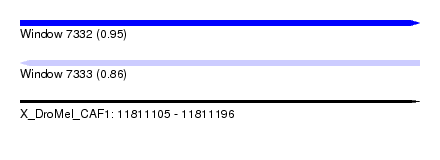
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,811,105 – 11,811,196 |
| Length | 91 |
| Max. P | 0.946047 |

| Location | 11,811,105 – 11,811,196 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 89.07 |
| Mean single sequence MFE | -21.28 |
| Consensus MFE | -16.28 |
| Energy contribution | -16.44 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11811105 91 + 22224390 GACAUGAUUGUCGCAGACAUUGAAAGCUCCUCCACGACAAUCAUGUGCCACAA-ACAAUCAUUCUAGUCAAA-AAGAGUACGUUUCAAACGAA .((((((((((((..((.............))..)))))))))))).......-......(((((.......-.))))).(((.....))).. ( -19.22) >DroSec_CAF1 14865 89 + 1 GACAUGAUUGUCGCAGACAUGGAUAGCUCCACCGCGACAAUCAUGUGCC-CAA-ACAACCAUUCUAGUCAAA-AAGAGUACGUUUCAAACGA- .(((((((((((((.(...((((....))))).)))))))))))))...-...-......(((((.......-.))))).(((.....))).- ( -26.20) >DroSim_CAF1 16738 91 + 1 GACAUGAUUGUCGCAGACAUGGAUAGCUCCGCCACGACAAUCAUGUGCCACAA-ACAACCAUUCUAGUCAAA-AAGAGUACGUUUCAAACGAA .((((((((((((..(....(((....))).)..)))))))))))).......-......(((((.......-.))))).(((.....))).. ( -21.60) >DroEre_CAF1 14698 88 + 1 GACAUGGUUGCCGCCGAUACGGAGUGCUC---CACGACAACCAUGUGCCACAA-ACAACCAUUCUAGUCAAA-AAGAGUACGUUUCAAACGAA .((((((((((((......))).(((...---)))..))))))))).......-......(((((.......-.))))).(((.....))).. ( -18.90) >DroYak_CAF1 11904 88 + 1 GACAUGAUUGUCGCAGCU--GGAAAGCUC---CACGACAACCAUGUGCCACAAAACAACCAUUCUAGUCAAAAAAGAGUACGUUUCAAACGAA .(((((.((((((.((((--....)))).---..)))))).)))))..............(((((.........))))).(((.....))).. ( -20.50) >consensus GACAUGAUUGUCGCAGACAUGGAAAGCUCC_CCACGACAAUCAUGUGCCACAA_ACAACCAUUCUAGUCAAA_AAGAGUACGUUUCAAACGAA .((((((((((((.....................))))))))))))..............(((((.........))))).(((.....))).. (-16.28 = -16.44 + 0.16)

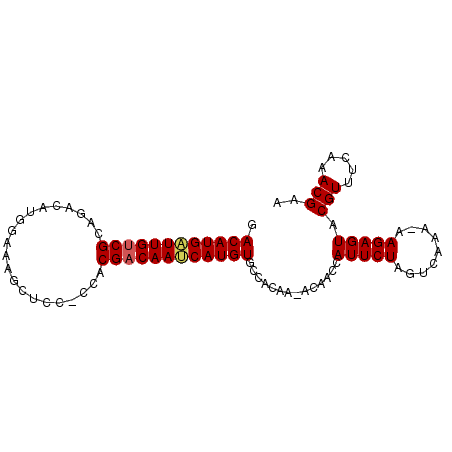

| Location | 11,811,105 – 11,811,196 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 89.07 |
| Mean single sequence MFE | -29.24 |
| Consensus MFE | -20.30 |
| Energy contribution | -20.02 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11811105 91 - 22224390 UUCGUUUGAAACGUACUCUU-UUUGACUAGAAUGAUUGU-UUGUGGCACAUGAUUGUCGUGGAGGAGCUUUCAAUGUCUGCGACAAUCAUGUC ...(((..(((((...(((.-.......))).....)))-))..)))((((((((((((..((.(.........).))..)))))))))))). ( -25.30) >DroSec_CAF1 14865 89 - 1 -UCGUUUGAAACGUACUCUU-UUUGACUAGAAUGGUUGU-UUG-GGCACAUGAUUGUCGCGGUGGAGCUAUCCAUGUCUGCGACAAUCAUGUC -..(((..((....(((.((-((.....)))).)))..)-)..-)))(((((((((((((((((((....))))...))))))))))))))). ( -34.20) >DroSim_CAF1 16738 91 - 1 UUCGUUUGAAACGUACUCUU-UUUGACUAGAAUGGUUGU-UUGUGGCACAUGAUUGUCGUGGCGGAGCUAUCCAUGUCUGCGACAAUCAUGUC ...(((..(((((.(((.((-((.....)))).))))))-))..)))(((((((((((((((((((....)))..))).))))))))))))). ( -29.60) >DroEre_CAF1 14698 88 - 1 UUCGUUUGAAACGUACUCUU-UUUGACUAGAAUGGUUGU-UUGUGGCACAUGGUUGUCGUG---GAGCACUCCGUAUCGGCGGCAACCAUGUC ...(((..(((((.(((.((-((.....)))).))))))-))..)))(((((((((((((.---((((.....)).)).))))))))))))). ( -29.60) >DroYak_CAF1 11904 88 - 1 UUCGUUUGAAACGUACUCUUUUUUGACUAGAAUGGUUGUUUUGUGGCACAUGGUUGUCGUG---GAGCUUUCC--AGCUGCGACAAUCAUGUC ...((((((((((.(((.((((......)))).)))))))))).)))(((((((((((((.---.((((....--))))))))))))))))). ( -27.50) >consensus UUCGUUUGAAACGUACUCUU_UUUGACUAGAAUGGUUGU_UUGUGGCACAUGAUUGUCGUGG_GGAGCUAUCCAUGUCUGCGACAAUCAUGUC .((((((....((..........))....))))))............(((((((((((((((...............))))))))))))))). (-20.30 = -20.02 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:40 2006