
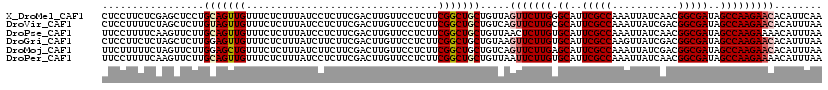
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,804,443 – 11,804,603 |
| Length | 160 |
| Max. P | 0.999989 |

| Location | 11,804,443 – 11,804,563 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -22.81 |
| Energy contribution | -23.42 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11804443 120 + 22224390 CUCCUUCUCGAGCUCCUGCAGUUGUUUCUCUUUAUCCUCUUCGACUUGUUCCUCUUCGGCUGCUGUUAGUUCUUGGGCAUUCGCCAAAUUAUCAACGGCGAUAGCCAAGAACACAUUCAA .........(((.....(((((((................................))))))).....((((((((.(..(((((...........)))))..)))))))))...))).. ( -27.55) >DroVir_CAF1 10613 120 + 1 CUCCUUUUCUAGCUCUUGUAGUUGUUUCUCUUUAUCCUCUUCGACUUGUUCCUCUUCGGCUGCUGUCAGUUCUUGCGCAUUCGCCAAAUUAUCGACGGCGAUAGCCAAGAACACAUUUAA .........(((((.....)))))..................(((..((.((.....))..)).))).(((((((.((..(((((...........)))))..)))))))))........ ( -25.60) >DroPse_CAF1 181 120 + 1 UUCCUUUUCAAGUUCUUGCAGUUGUUUCUCUUUAUCCUCUUCGACUUGUUCCUCUUCGGCUGCUGUUAACUCUUGUGCAUUCGCCAAAUUAUCAACGGCGAUAGCCAAGAAAACAUUUAA .................(((((((................................)))))))((((...(((((.((..(((((...........)))))..))))))).))))..... ( -23.15) >DroGri_CAF1 10143 120 + 1 CUCCUUCUCUAGCUCUUGGAGUUGUUUCUCUUUAUCUUCUUCGACUUGUUCCUCUUCGGCUGCUGUAAGUUCUUGUGCAUUCGCCAAGUUAUCGACGGCGAUAGCCAAGAACACAUUUAA .........((((..((((((..(..........................)..))))))..))))...(((((((.((..(((((...........)))))..)))))))))........ ( -26.27) >DroMoj_CAF1 8922 120 + 1 UUCUUUUUCUAGUUCUUGGAGCUGUUUCUCUUUAUCUUCUUCGACUUGUUCCUCUUCGGCUGCUGUCAGUUCUUGAGCAUUCGCCAAAUUAUCGACGGCGAUAGCCAAGAACACAUUUAA ...........(((((((((((.(((................)))..))))).....((((((((((.((..(((.((....)))))...)).))))))...))))))))))........ ( -29.39) >DroPer_CAF1 6247 120 + 1 UUCCUUUUCAAGUUCUUGCAGUUGUUUCUCUUUAUCCUCUUCGACUUGUUCCUCUUCGGCUGCUGUUAAUUCUUGUGCAUUCGCCAAAUUAUCAACGGCGAUAGCCAAGAAAACAUUUAA .................(((((((................................)))))))((((..((((((.((..(((((...........)))))..))))))))))))..... ( -24.35) >consensus CUCCUUUUCUAGCUCUUGCAGUUGUUUCUCUUUAUCCUCUUCGACUUGUUCCUCUUCGGCUGCUGUUAGUUCUUGUGCAUUCGCCAAAUUAUCAACGGCGAUAGCCAAGAACACAUUUAA .................(((((((................................))))))).....(((((((.((..(((((...........)))))..)))))))))........ (-22.81 = -23.42 + 0.61)
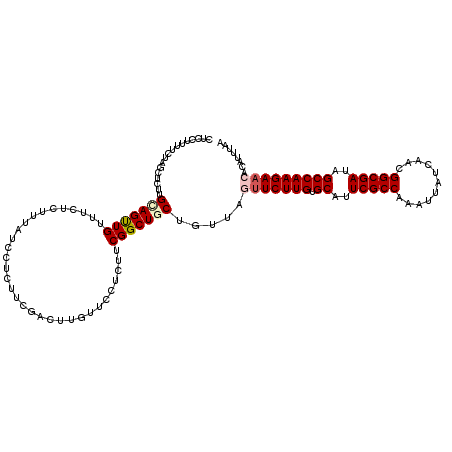

| Location | 11,804,483 – 11,804,603 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -32.71 |
| Energy contribution | -32.98 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.70 |
| SVM RNA-class probability | 0.999941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11804483 120 + 22224390 UCGACUUGUUCCUCUUCGGCUGCUGUUAGUUCUUGGGCAUUCGCCAAAUUAUCAACGGCGAUAGCCAAGAACACAUUCAAUAGCGUGUAAUUUCCGAAGAGUACCAAAACGAUAAAGUAU (((..(((...((((((((.........((((((((.(..(((((...........)))))..)))))))))((((.(....).)))).....))))))))...)))..)))........ ( -36.30) >DroVir_CAF1 10653 120 + 1 UCGACUUGUUCCUCUUCGGCUGCUGUCAGUUCUUGCGCAUUCGCCAAAUUAUCGACGGCGAUAGCCAAGAACACAUUUAAAAGCGUAUAAUUACCGAAGAGUACCAAAACGAUAAAGUAU (((..(((...((((((((((..(((..(((((((.((..(((((...........)))))..))))))))))))......)))(((....))))))))))...)))..)))........ ( -34.90) >DroPse_CAF1 221 120 + 1 UCGACUUGUUCCUCUUCGGCUGCUGUUAACUCUUGUGCAUUCGCCAAAUUAUCAACGGCGAUAGCCAAGAAAACAUUUAAGAGCGUGUAAUUACCGAAGAGUACCAAAACGAUAAAGUAU (((..(((...(((((((((((((.((((.(((((.((..(((((...........)))))..)))))))......)))).)))).)......))))))))...)))..)))........ ( -32.60) >DroGri_CAF1 10183 120 + 1 UCGACUUGUUCCUCUUCGGCUGCUGUAAGUUCUUGUGCAUUCGCCAAGUUAUCGACGGCGAUAGCCAAGAACACAUUUAAAAGCGUAUAAUUACCGAAGAGUACCAAAACGAUAAAGUAU (((..(((...((((((((((...((..(((((((.((..(((((...........)))))..)))))))))..)).....)))(((....))))))))))...)))..)))........ ( -35.00) >DroMoj_CAF1 8962 120 + 1 UCGACUUGUUCCUCUUCGGCUGCUGUCAGUUCUUGAGCAUUCGCCAAAUUAUCGACGGCGAUAGCCAAGAACACAUUUAAAAGCGUAUAAUUACCGAAGAGUACCAAAACGAUAAAGUAU (((..(((...((((((((((..(((..(((((((.((..(((((...........)))))..))))))))))))......)))(((....))))))))))...)))..)))........ ( -34.90) >DroPer_CAF1 6287 120 + 1 UCGACUUGUUCCUCUUCGGCUGCUGUUAAUUCUUGUGCAUUCGCCAAAUUAUCAACGGCGAUAGCCAAGAAAACAUUUAAGAGCGUGUAAUUACCGAAGAGUACCAAAACGAUAAAGUAU (((..(((...(((((((((((((.((((((((((.((..(((((...........)))))..)))))))).....)))).)))).)......))))))))...)))..)))........ ( -34.60) >consensus UCGACUUGUUCCUCUUCGGCUGCUGUUAGUUCUUGUGCAUUCGCCAAAUUAUCAACGGCGAUAGCCAAGAACACAUUUAAAAGCGUAUAAUUACCGAAGAGUACCAAAACGAUAAAGUAU (((..(((...((((((((((..((...(((((((.((..(((((...........)))))..))))))))).....))..)))(((....))))))))))...)))..)))........ (-32.71 = -32.98 + 0.28)


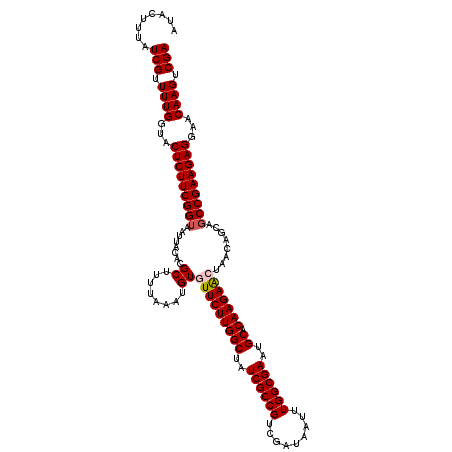
| Location | 11,804,483 – 11,804,603 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -39.70 |
| Consensus MFE | -38.31 |
| Energy contribution | -38.67 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.96 |
| SVM decision value | 5.52 |
| SVM RNA-class probability | 0.999989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11804483 120 - 22224390 AUACUUUAUCGUUUUGGUACUCUUCGGAAAUUACACGCUAUUGAAUGUGUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCCCAAGAACUAACAGCAGCCGAAGAGGAACAAGUCGA ........(((.((((...((((((((.....(((..(....)..)))(((((((((..((((((.........))))))..).)))))))).........))))))))...)))).))) ( -40.30) >DroVir_CAF1 10653 120 - 1 AUACUUUAUCGUUUUGGUACUCUUCGGUAAUUAUACGCUUUUAAAUGUGUUCUUGGCUAUCGCCGUCGAUAAUUUGGCGAAUGCGCAAGAACUGACAGCAGCCGAAGAGGAACAAGUCGA ........(((.((((...(((((((((........((........))(((((((((..((((((.........))))))..)).)))))))........)))))))))...)))).))) ( -40.80) >DroPse_CAF1 221 120 - 1 AUACUUUAUCGUUUUGGUACUCUUCGGUAAUUACACGCUCUUAAAUGUUUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCACAAGAGUUAACAGCAGCCGAAGAGGAACAAGUCGA ........(((.((((...(((((((((........(((.((((.....((((((((..((((((.........))))))..)).)))))))))).))).)))))))))...)))).))) ( -37.60) >DroGri_CAF1 10183 120 - 1 AUACUUUAUCGUUUUGGUACUCUUCGGUAAUUAUACGCUUUUAAAUGUGUUCUUGGCUAUCGCCGUCGAUAACUUGGCGAAUGCACAAGAACUUACAGCAGCCGAAGAGGAACAAGUCGA ........(((.((((...(((((((((........((........))(((((((((..((((((.........))))))..)).)))))))........)))))))))...)))).))) ( -40.80) >DroMoj_CAF1 8962 120 - 1 AUACUUUAUCGUUUUGGUACUCUUCGGUAAUUAUACGCUUUUAAAUGUGUUCUUGGCUAUCGCCGUCGAUAAUUUGGCGAAUGCUCAAGAACUGACAGCAGCCGAAGAGGAACAAGUCGA ........(((.((((...(((((((((........((........))(((((((((..((((((.........))))))..)).)))))))........)))))))))...)))).))) ( -40.80) >DroPer_CAF1 6287 120 - 1 AUACUUUAUCGUUUUGGUACUCUUCGGUAAUUACACGCUCUUAAAUGUUUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCACAAGAAUUAACAGCAGCCGAAGAGGAACAAGUCGA ........(((.((((...(((((((((........(((.((((.....((((((((..((((((.........))))))..)).)))))))))).))).)))))))))...)))).))) ( -37.90) >consensus AUACUUUAUCGUUUUGGUACUCUUCGGUAAUUACACGCUUUUAAAUGUGUUCUUGGCUAUCGCCGUCGAUAAUUUGGCGAAUGCACAAGAACUAACAGCAGCCGAAGAGGAACAAGUCGA ........(((.((((...(((((((((........((........))(((((((((..((((((.........))))))..)).)))))))........)))))))))...)))).))) (-38.31 = -38.67 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:38 2006