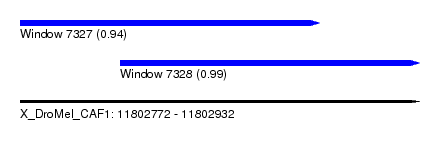
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,802,772 – 11,802,932 |
| Length | 160 |
| Max. P | 0.993153 |

| Location | 11,802,772 – 11,802,892 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -28.25 |
| Energy contribution | -28.86 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
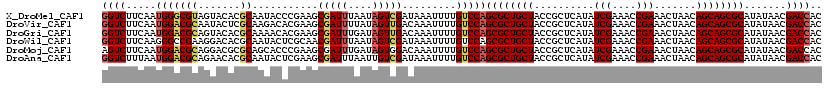
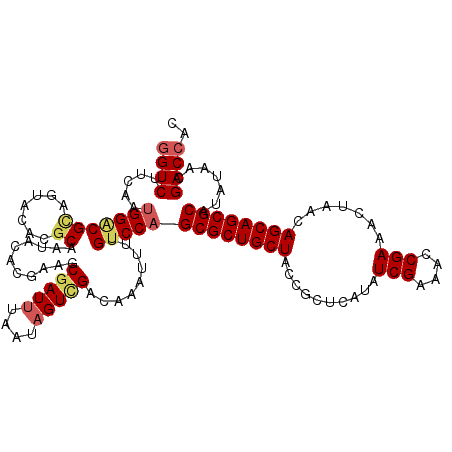
>X_DroMel_CAF1 11802772 120 + 22224390 GGUCUUCAAUGGGCGUAGUACACGCAAUACCCGAAGCGAUUUAAUAGUCGAUAAAUUUUGUCCAGCGCUGCUACCGCUCAUAUCGAAACCGAAACUAACAGCAGCGCAUAUAACGACCAC ((((.....(((((((.....))))...........(((((....)))))...........)))((((((((..........(((....))).......)))))))).......)))).. ( -32.53) >DroVir_CAF1 8489 120 + 1 GGUCUUCAAUGGACGCAAUACUCGCAAGACACGAAGCGAUUUUAUAGUUGACAAAUUUUGUCCAGCGCUGCUACCGCUCAUAUCGAAACCGAAACUAACAGCAGCGCAUAUAACGACCAC ((((.....((((((.(((..((....))......((((((....))))).)..))).))))))((((((((..........(((....))).......)))))))).......)))).. ( -33.73) >DroGri_CAF1 8310 120 + 1 GGUCUUCAAUGGACGCAGUACACGCAAAACACGAAGCGAUUUGAUAGUUGACAAAUUUUGUCCAGCGCUGCUACCGCUCAUAUCGAAACCGAAACUAACAGCAGCGCAUAUAACGACCAC ((((.....(((((........(((..........)))(((((........)))))...)))))((((((((..........(((....))).......)))))))).......)))).. ( -32.63) >DroWil_CAF1 8903 120 + 1 GGUCUUCAAGGGCCGAAGGACACGCAAUACUCGCAACGAUUUAAUAGUCGAUAAAUUUUGUCCAGCGCUGCUACCGCUCAUAUCGAAACCGAAACUAACAGCAGCGCAUAUAACGACCAC ((((.............(((((.((.......))..(((((....)))))........))))).((((((((..........(((....))).......)))))))).......)))).. ( -34.13) >DroMoj_CAF1 7103 120 + 1 AGUCUUCAAUGGACGCAGGACGCGCAGCACCCGAAGCGAUUUGAUAGUGGACAAAUUUUGUCCAGCGCUGCUACCGCUCAUAUCGAAACCGAAACUAACAGCAGCGCAUAUAACGACCAC .((((.....))))...((.(((((..(....)..))).........((((((.....))))))((((((((..........(((....))).......))))))))......)).)).. ( -31.93) >DroAna_CAF1 5953 120 + 1 GGUCUUUAAUGGACGCAGAACACGCAAUACUCGAAGCGAUUUAAUUGUCGAUAAAUUUUGUCCAGCGCUGCUACCGCUCAUAUCGAAACCGAAACUAACAGCAGCGCAUAUAACGACCAC ((((.(((.(((((((.......))...........((((......)))).........)))))((((((((..........(((....))).......))))))))...))).)))).. ( -33.83) >consensus GGUCUUCAAUGGACGCAGUACACGCAAUACACGAAGCGAUUUAAUAGUCGACAAAUUUUGUCCAGCGCUGCUACCGCUCAUAUCGAAACCGAAACUAACAGCAGCGCAUAUAACGACCAC ((((.....(((((((.......))...........(((((....))))).........)))))((((((((..........(((....))).......)))))))).......)))).. (-28.25 = -28.86 + 0.61)

| Location | 11,802,812 – 11,802,932 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.44 |
| Mean single sequence MFE | -22.76 |
| Consensus MFE | -21.41 |
| Energy contribution | -21.33 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
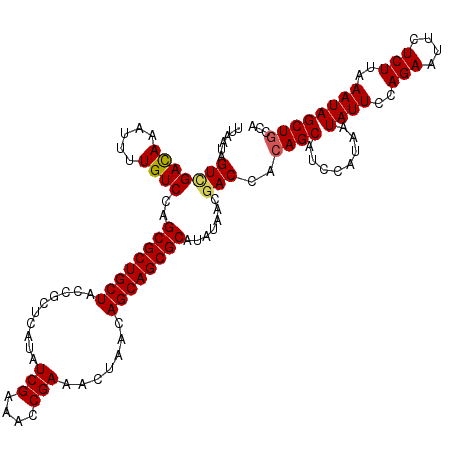
>X_DroMel_CAF1 11802812 120 + 22224390 UUAAUAGUCGAUAAAUUUUGUCCAGCGCUGCUACCGCUCAUAUCGAAACCGAAACUAACAGCAGCGCAUAUAACGACCACAGCAUCCAUAAUAUUCCAGAAUUCUCUUAAAUAGCUGCCA ......(((((((.....))))..((((((((..........(((....))).......)))))))).......)))..((((........((((..(((....)))..))))))))... ( -22.83) >DroVir_CAF1 8529 120 + 1 UUUAUAGUUGACAAAUUUUGUCCAGCGCUGCUACCGCUCAUAUCGAAACCGAAACUAACAGCAGCGCAUAUAACGACCACAGCAUCCAUAAUAUUCCAGAAUUCUCUUAAAUAGCUGCCA .(((((...((((.....))))..((((((((..........(((....))).......)))))))).)))))......((((........((((..(((....)))..))))))))... ( -22.23) >DroGri_CAF1 8350 120 + 1 UUGAUAGUUGACAAAUUUUGUCCAGCGCUGCUACCGCUCAUAUCGAAACCGAAACUAACAGCAGCGCAUAUAACGACCACAGCAUCCAUAAUAUUCCAGAAUUCUCUUAAAUAGCUGCCA ......(((((((.....))))..((((((((..........(((....))).......))))))))....))).....((((........((((..(((....)))..))))))))... ( -22.23) >DroMoj_CAF1 7143 120 + 1 UUGAUAGUGGACAAAUUUUGUCCAGCGCUGCUACCGCUCAUAUCGAAACCGAAACUAACAGCAGCGCAUAUAACGACCACAGCAUCCAUAAUAUUCCAGAAUUCUCUUAAAUAGCUGCCA .......((((((.....))))))((((((((..........(((....))).......))))))))............((((........((((..(((....)))..))))))))... ( -26.53) >DroAna_CAF1 5993 120 + 1 UUAAUUGUCGAUAAAUUUUGUCCAGCGCUGCUACCGCUCAUAUCGAAACCGAAACUAACAGCAGCGCAUAUAACGACCACAGCAUCCAUAAUAUUCCAGAAUUCUCUUAAAUAGCUUCCC ......(((((((.....))))..((((((((..........(((....))).......)))))))).......)))..............((((..(((....)))..))))....... ( -19.93) >DroPer_CAF1 4540 120 + 1 UUAAUAGUCGAUAAAUUUUGUCCAGCGCUGCUACCGCUCAUAUCGAAACCGAAACUAACAGCAGCGCAUAUAACGACCACAGCAUCCAUAAUAUUCCAGAAUUCUCUUAAAUAGCUGCCA ......(((((((.....))))..((((((((..........(((....))).......)))))))).......)))..((((........((((..(((....)))..))))))))... ( -22.83) >consensus UUAAUAGUCGACAAAUUUUGUCCAGCGCUGCUACCGCUCAUAUCGAAACCGAAACUAACAGCAGCGCAUAUAACGACCACAGCAUCCAUAAUAUUCCAGAAUUCUCUUAAAUAGCUGCCA ......(((((((.....))))..((((((((..........(((....))).......)))))))).......)))..((((........((((..(((....)))..))))))))... (-21.41 = -21.33 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:35 2006