
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,797,171 – 11,797,266 |
| Length | 95 |
| Max. P | 0.925693 |

| Location | 11,797,171 – 11,797,266 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.31 |
| Mean single sequence MFE | -20.42 |
| Consensus MFE | -12.92 |
| Energy contribution | -12.47 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
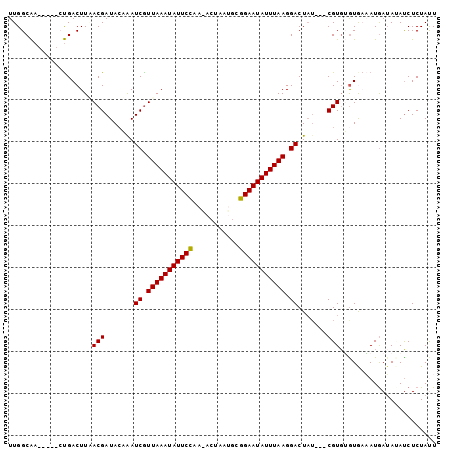
>X_DroMel_CAF1 11797171 95 + 22224390 UUGGCAA-----CUGACUUUACGAUACAAAUCAUUAAAUAUUCCCAGACUAAUGCGGAAUAUUUAAGGACU--UAUCGUUUGUGAAAUGAUAUAUCUCUAUU ..(....-----)(.((...((((((....((.(((((((((((((......)).))))))))))).))..--))))))..)).)................. ( -19.60) >DroVir_CAF1 3254 92 + 1 UUGGCGAAAAUAGCGACUUGACGAUACCAAUCGUUAAAUAUUCA-------UCACUGAAUAUUUAAGGACUAU---CGUGGGUAUUAUAAAAAAUCUCUACU ...((.......)).((((.((((((....((.(((((((((((-------....))))))))))).)).)))---)))))))................... ( -19.70) >DroGri_CAF1 2514 92 + 1 UUGGCAACAAUGGUGACUUGACGAUACCAAUCGUUAAAUAUUCA-------AAACUGAAUAUUUAAGGACUAU---CGUGAAUAUUAUGAUAAAUCUCUACU (((....)))(((((.(.....).))))).((.(((((((((((-------....))))))))))).)).(((---(((((...)))))))).......... ( -21.90) >DroSec_CAF1 992 97 + 1 UUGGCAA-----CUGACUUAACGAUACAGAUCGUUAAAUAUUCCAAAACUAAUGCGGAAUAUUUAAGGACUAUCAUCGUGUGUGAAAUGAUAUAUCUCUAUU ..(....-----)(.((...(((((..((.((.(((((((((((...........))))))))))).))))...)))))..)).)................. ( -19.60) >DroSim_CAF1 987 94 + 1 UUGGCAA-----CUGACUUAACGAUACAGAUCGUUAAAUAUUCCAAAACUAAUGCGGAAUAUUUAAGGACUAU---CGUGUGUGAAAUGAUAUAUCUCUAUU ..(....-----)(.((...(((((..((.((.(((((((((((...........))))))))))).))))))---)))..)).)................. ( -19.50) >DroEre_CAF1 997 94 + 1 UUGGCAA-----CUGACUUUACGAUAAAAAUCGUUAAAUAUUCCAAGACUACUGGGGAAUAUUUAAGGACUAU---CGUGUGUGAAAUGAUACACCUCUAUU ..(....-----).......((((((....((.(((((((((((.((....))..))))))))))).)).)))---)))(((((......)))))....... ( -22.20) >consensus UUGGCAA_____CUGACUUAACGAUACAAAUCGUUAAAUAUUCCAA_ACUAAUGCGGAAUAUUUAAGGACUAU___CGUGUGUGAAAUGAUAUAUCUCUAUU ....................(((.......((.(((((((((((...........))))))))))).)).......)))....................... (-12.92 = -12.47 + -0.44)


| Location | 11,797,171 – 11,797,266 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 78.31 |
| Mean single sequence MFE | -20.23 |
| Consensus MFE | -12.98 |
| Energy contribution | -12.87 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
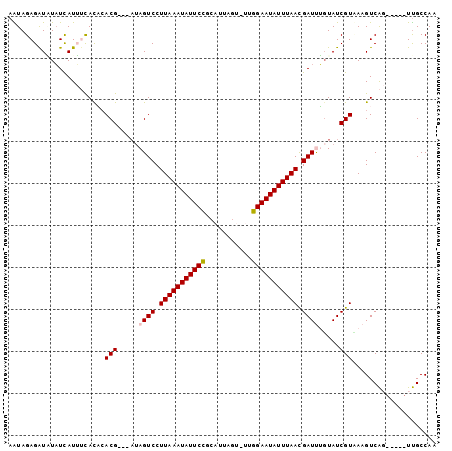
>X_DroMel_CAF1 11797171 95 - 22224390 AAUAGAGAUAUAUCAUUUCACAAACGAUA--AGUCCUUAAAUAUUCCGCAUUAGUCUGGGAAUAUUUAAUGAUUUGUAUCGUAAAGUCAG-----UUGCCAA ....(((((.....)))))....((((((--.(((.(((((((((((.((......))))))))))))).)))...))))))........-----....... ( -22.20) >DroVir_CAF1 3254 92 - 1 AGUAGAGAUUUUUUAUAAUACCCACG---AUAGUCCUUAAAUAUUCAGUGA-------UGAAUAUUUAACGAUUGGUAUCGUCAAGUCGCUAUUUUCGCCAA (((((.(((((...(..(((((....---..((((.(((((((((((....-------))))))))))).)))))))))..).))))).)))))........ ( -22.60) >DroGri_CAF1 2514 92 - 1 AGUAGAGAUUUAUCAUAAUAUUCACG---AUAGUCCUUAAAUAUUCAGUUU-------UGAAUAUUUAACGAUUGGUAUCGUCAAGUCACCAUUGUUGCCAA .(((((......)).........(((---((((((.((((((((((.....-------.)))))))))).))))(((..(.....)..)))))))))))... ( -15.80) >DroSec_CAF1 992 97 - 1 AAUAGAGAUAUAUCAUUUCACACACGAUGAUAGUCCUUAAAUAUUCCGCAUUAGUUUUGGAAUAUUUAACGAUCUGUAUCGUUAAGUCAG-----UUGCCAA ....(((((.....)))))....(((((.((((((.((((((((((((.........)))))))))))).)).)))))))))........-----....... ( -21.00) >DroSim_CAF1 987 94 - 1 AAUAGAGAUAUAUCAUUUCACACACG---AUAGUCCUUAAAUAUUCCGCAUUAGUUUUGGAAUAUUUAACGAUCUGUAUCGUUAAGUCAG-----UUGCCAA ....(((((.....)))))....(((---((((((.((((((((((((.........)))))))))))).)))...))))))........-----....... ( -19.10) >DroEre_CAF1 997 94 - 1 AAUAGAGGUGUAUCAUUUCACACACG---AUAGUCCUUAAAUAUUCCCCAGUAGUCUUGGAAUAUUUAACGAUUUUUAUCGUAAAGUCAG-----UUGCCAA ....((((((...))))))....(((---((((((.(((((((((((..((....)).))))))))))).)))...))))))........-----....... ( -20.70) >consensus AAUAGAGAUAUAUCAUUUCACACACG___AUAGUCCUUAAAUAUUCCGCAUUAGU_UUGGAAUAUUUAACGAUUUGUAUCGUAAAGUCAG_____UUGCCAA .......................(((.....((((.(((((((((((...........))))))))))).)))).....))).................... (-12.98 = -12.87 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:31 2006