
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,789,518 – 11,789,661 |
| Length | 143 |
| Max. P | 0.999920 |

| Location | 11,789,518 – 11,789,621 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 90.95 |
| Mean single sequence MFE | -40.80 |
| Consensus MFE | -35.68 |
| Energy contribution | -36.96 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.97 |
| Structure conservation index | 0.87 |
| SVM decision value | 4.56 |
| SVM RNA-class probability | 0.999920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11789518 103 + 22224390 AA--CCCGGUAUGCGUCCGAUCAUGCGCGAUG--UGAAUGUGGCAUUAAGUGGCAGUUAAAAU-GUAACCGCCACAUCAACCGCUGAUUUAAGGACGCAUACCGCCUC ..--..(((((((((((((((((.((((.((.--.....)).)).....(((((.((((....-.)))).))))).......)))))))...)))))))))))).... ( -43.80) >DroSec_CAF1 19129 103 + 1 AA--CCAGGGUUGCGUCCGAUCAUGCGCGAUG--UGAAUGUGGCAUUAAGUGGCGGUUAAAAU-GUAACCGCCACAUCAACCGCUGAUUUAAGGACGCAUACCGCCUC ..--...((..((((((((((((.((((.((.--.....)).)).....((((((((((....-.)))))))))).......)))))))...)))))))..))..... ( -43.30) >DroSim_CAF1 19313 103 + 1 AU--CCCGGUAUGCGUCCGAUCAUGCGCGAUG--UGAAUGUGGCAUUAAGUGGCGGUUAAAAU-GUAACCGCCACAUCAACCGCUGAUUUAAGGACGCAUACCGCCUC ..--..(((((((((((((((((.((((.((.--.....)).)).....((((((((((....-.)))))))))).......)))))))...)))))))))))).... ( -49.90) >DroEre_CAF1 18921 103 + 1 AA--CCCGGUUUGCGUCCGAUCAUGCGCGAUG--UGAAUGUGGUAUUAUGUGCCUGUUAAAUU-UUAAUCGACACAUCAACCGCUGAUUUAUGGACGCAUACCGCCUC ..--..((((.((((((((((((.(((.((((--((.....(((((...))))).((((....-.))))...))))))...))))))))...))))))).)))).... ( -35.70) >DroYak_CAF1 19569 108 + 1 AAUCCCCGCUUUGCGACCGAUCAUGCGCGAUGUGUGAAUGUGGCAUUAAGUGGCGGUUAAAUUGUUAACCGACACUUCAACCGCUGAUUUAAGGACGCAUACCGCCUC .......((..((((.(((((((.((((.((((....)))).))...(((((.(((((((....))))))).))))).....)))))))...)).))))....))... ( -31.30) >consensus AA__CCCGGUUUGCGUCCGAUCAUGCGCGAUG__UGAAUGUGGCAUUAAGUGGCGGUUAAAAU_GUAACCGCCACAUCAACCGCUGAUUUAAGGACGCAUACCGCCUC ......((((.((((((((((((.((((.((........)).)).....((((((((((......)))))))))).......)))))))...))))))).)))).... (-35.68 = -36.96 + 1.28)


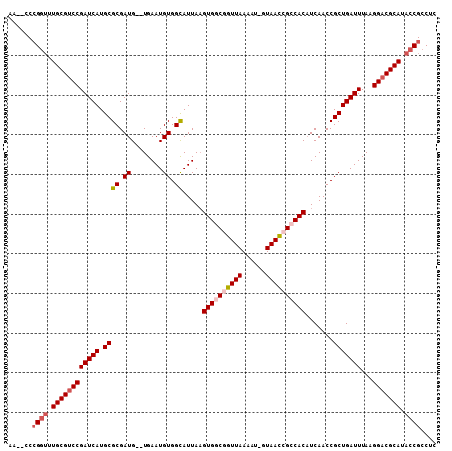
| Location | 11,789,518 – 11,789,621 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 90.95 |
| Mean single sequence MFE | -39.18 |
| Consensus MFE | -33.38 |
| Energy contribution | -34.26 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.93 |
| SVM RNA-class probability | 0.999712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11789518 103 - 22224390 GAGGCGGUAUGCGUCCUUAAAUCAGCGGUUGAUGUGGCGGUUAC-AUUUUAACUGCCACUUAAUGCCACAUUCA--CAUCGCGCAUGAUCGGACGCAUACCGGG--UU ....((((((((((((....((((((.(((((.((((((((((.-....)))))))))))))))((........--....)))).)))).))))))))))))..--.. ( -44.60) >DroSec_CAF1 19129 103 - 1 GAGGCGGUAUGCGUCCUUAAAUCAGCGGUUGAUGUGGCGGUUAC-AUUUUAACCGCCACUUAAUGCCACAUUCA--CAUCGCGCAUGAUCGGACGCAACCCUGG--UU ..((.(((.(((((((....((((((.(((((.((((((((((.-....)))))))))))))))((........--....)))).)))).))))))))))))..--.. ( -39.70) >DroSim_CAF1 19313 103 - 1 GAGGCGGUAUGCGUCCUUAAAUCAGCGGUUGAUGUGGCGGUUAC-AUUUUAACCGCCACUUAAUGCCACAUUCA--CAUCGCGCAUGAUCGGACGCAUACCGGG--AU ....((((((((((((....((((((.(((((.((((((((((.-....)))))))))))))))((........--....)))).)))).))))))))))))..--.. ( -46.80) >DroEre_CAF1 18921 103 - 1 GAGGCGGUAUGCGUCCAUAAAUCAGCGGUUGAUGUGUCGAUUAA-AAUUUAACAGGCACAUAAUACCACAUUCA--CAUCGCGCAUGAUCGGACGCAAACCGGG--UU ....((((.(((((((....(((((((((..(((((((..(((.-....)))..)))))))...))).....(.--....).)).)))).))))))).))))..--.. ( -33.80) >DroYak_CAF1 19569 108 - 1 GAGGCGGUAUGCGUCCUUAAAUCAGCGGUUGAAGUGUCGGUUAACAAUUUAACCGCCACUUAAUGCCACAUUCACACAUCGCGCAUGAUCGGUCGCAAAGCGGGGAUU ....((.(.((((.((....(((((((((..(((((.(((((((....))))))).)))))...))).......(.....).)).)))).)).)))).).))...... ( -31.00) >consensus GAGGCGGUAUGCGUCCUUAAAUCAGCGGUUGAUGUGGCGGUUAC_AUUUUAACCGCCACUUAAUGCCACAUUCA__CAUCGCGCAUGAUCGGACGCAAACCGGG__UU ....((((.(((((((....(((((((((....((((((((((......)))))))))).....))).....(.......).)).)))).))))))).))))...... (-33.38 = -34.26 + 0.88)


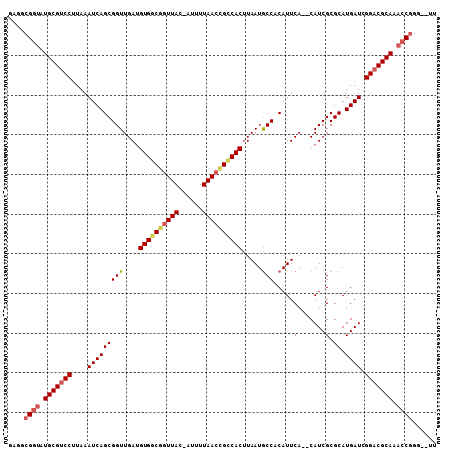
| Location | 11,789,551 – 11,789,661 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 91.76 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -26.74 |
| Energy contribution | -27.42 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11789551 110 + 22224390 AUGUGGCAUUAAGUGGCAGUUAAAAU-GUAACCGCCACAUCAACCGCUGAUUUAAGGACGCAUACCGCCUCACCGUGGAAUCUGCAUUGAUAAAUCACAUUCGACUCGACA ((((((......(((((.((((....-.)))).)))))(((((..((.(((((..((..((.....))....))...))))).)).)))))...))))))........... ( -27.90) >DroSec_CAF1 19162 110 + 1 AUGUGGCAUUAAGUGGCGGUUAAAAU-GUAACCGCCACAUCAACCGCUGAUUUAAGGACGCAUACCGCCUCACCGUAGGAUCUGCAUUGAUAAAUCACAUUCGACUCGACA ((((((......((((((((((....-.))))))))))(((((..((.(((((..((..((.....))....))...))))).)).)))))...))))))........... ( -32.50) >DroSim_CAF1 19346 110 + 1 AUGUGGCAUUAAGUGGCGGUUAAAAU-GUAACCGCCACAUCAACCGCUGAUUUAAGGACGCAUACCGCCUCACCGUGGGAUCUGCAUUGAUAAAUCACAUUCGACUCGACA ..((((......((((((((((....-.)))))))))).....))))(((((((..((.(((..((((......))))....))).))..))))))).............. ( -33.60) >DroEre_CAF1 18954 110 + 1 AUGUGGUAUUAUGUGCCUGUUAAAUU-UUAAUCGACACAUCAACCGCUGAUUUAUGGACGCAUACCGCCUCACCGUGGGAUCUGCAUUGAUAAAUCGCACUCGACUCGGCG ..(((((...(((((.(.((((....-.)))).).)))))..))))).(((((((.((.(((..((((......))))....))).)).)))))))((.(.......))). ( -27.20) >DroYak_CAF1 19606 111 + 1 AUGUGGCAUUAAGUGGCGGUUAAAUUGUUAACCGACACUUCAACCGCUGAUUUAAGGACGCAUACCGCCUCACCGUGGGUUCUGCAUUGUUAAAUCACGUUCGACUCGACG ..((((....(((((.(((((((....))))))).)))))...))))((((((((.((.(((..((((......))))....))).)).))))))))((((......)))) ( -34.60) >consensus AUGUGGCAUUAAGUGGCGGUUAAAAU_GUAACCGCCACAUCAACCGCUGAUUUAAGGACGCAUACCGCCUCACCGUGGGAUCUGCAUUGAUAAAUCACAUUCGACUCGACA ..((((......((((((((((......)))))))))).....))))(((((((..((.(((..((((......))))....))).))..))))))).............. (-26.74 = -27.42 + 0.68)


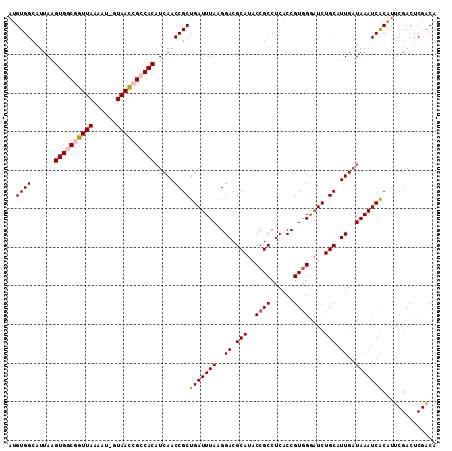
| Location | 11,789,551 – 11,789,661 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 91.76 |
| Mean single sequence MFE | -33.94 |
| Consensus MFE | -30.76 |
| Energy contribution | -30.76 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11789551 110 - 22224390 UGUCGAGUCGAAUGUGAUUUAUCAAUGCAGAUUCCACGGUGAGGCGGUAUGCGUCCUUAAAUCAGCGGUUGAUGUGGCGGUUAC-AUUUUAACUGCCACUUAAUGCCACAU .((.(((((.....(((....))).....))))).))(((((((((.....)).)))).........(((((.((((((((((.-....)))))))))))))))))).... ( -33.30) >DroSec_CAF1 19162 110 - 1 UGUCGAGUCGAAUGUGAUUUAUCAAUGCAGAUCCUACGGUGAGGCGGUAUGCGUCCUUAAAUCAGCGGUUGAUGUGGCGGUUAC-AUUUUAACCGCCACUUAAUGCCACAU .(((((.(((....(((((((...(((((...((..(.....)..))..)))))...))))))).))))))))((((((((((.-....))))))))))............ ( -35.00) >DroSim_CAF1 19346 110 - 1 UGUCGAGUCGAAUGUGAUUUAUCAAUGCAGAUCCCACGGUGAGGCGGUAUGCGUCCUUAAAUCAGCGGUUGAUGUGGCGGUUAC-AUUUUAACCGCCACUUAAUGCCACAU .(((((.(((....(((((((...(((((.(((((.......)).))).)))))...))))))).))))))))((((((((((.-....))))))))))............ ( -36.30) >DroEre_CAF1 18954 110 - 1 CGCCGAGUCGAGUGCGAUUUAUCAAUGCAGAUCCCACGGUGAGGCGGUAUGCGUCCAUAAAUCAGCGGUUGAUGUGUCGAUUAA-AAUUUAACAGGCACAUAAUACCACAU .(((..(((((.((((((((((..(((((.(((((.......)).))).)))))..))))))).))).))))).(((..(....-...)..)))))).............. ( -30.60) >DroYak_CAF1 19606 111 - 1 CGUCGAGUCGAACGUGAUUUAACAAUGCAGAACCCACGGUGAGGCGGUAUGCGUCCUUAAAUCAGCGGUUGAAGUGUCGGUUAACAAUUUAACCGCCACUUAAUGCCACAU (((((...)).)))((((((((..(((((..(((..(.....)..))).)))))..))))))))(.(((..(((((.(((((((....))))))).)))))...))).).. ( -34.50) >consensus UGUCGAGUCGAAUGUGAUUUAUCAAUGCAGAUCCCACGGUGAGGCGGUAUGCGUCCUUAAAUCAGCGGUUGAUGUGGCGGUUAC_AUUUUAACCGCCACUUAAUGCCACAU ......(((((.(((((((((...(((((.(((((.......)).))).)))))...)))))).))).)))))((((((((((......))))))))))............ (-30.76 = -30.76 + -0.00)

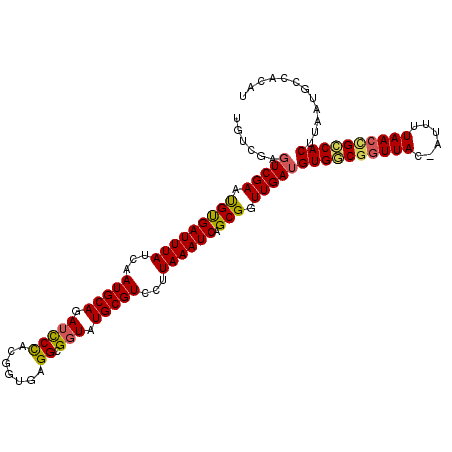

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:30 2006