
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,777,679 – 11,777,784 |
| Length | 105 |
| Max. P | 0.846450 |

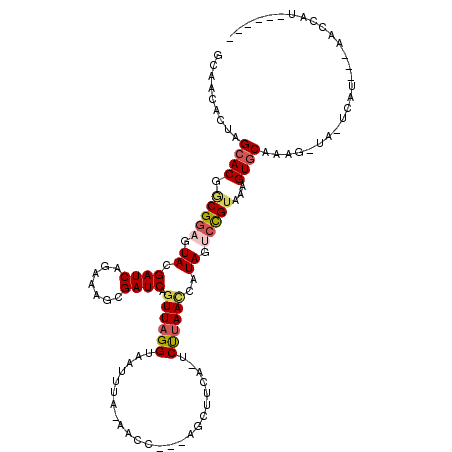
| Location | 11,777,679 – 11,777,784 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.47 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -15.81 |
| Energy contribution | -16.48 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11777679 105 - 22224390 GCAACACUAGCACGGCGGAGUACGAUCAGAAAGCGAUCAGUUAGGUAAUUUAAAACC---AGCUUCAAUCUUAACCAUAGUCCGUAAAGUGCAAAG-UA-UCAUAU-AAUC-------- .........((((.(((((.((.((((.......))))((((.(((........)))---)))).............)).)))))...))))....-..-......-....-------- ( -23.70) >DroEre_CAF1 7458 102 - 1 GCAACACUAGCACGGCGGAGUACGAUCAGAAAGCGAUCAGUUAGGUAAUUUA--ACC---AGCUUCAAUCUUAA-CAUAGUCCGUAAAGUGCAAAG-AA-UCAU---AACCAU------ .........((((.(((((.((.((((.......))))((((.(((......--)))---))))..........-..)).)))))...))))....-..-....---......------ ( -23.90) >DroWil_CAF1 13579 112 - 1 GCAACACUAGCACGACGGAGUACGAUCAGAAAGCGAUCAGUUAGGUAAUUUACAAACAAAAGCUUCA-UCCUAAUUAUAACCCGUAAAGUCCAAAAGUACUUACUUACUUACU------ .........(.((.((((.((..((((.......))))(((((((......................-.)))))))...))))))...)).)..(((((......)))))...------ ( -16.80) >DroYak_CAF1 7537 104 - 1 GCAACACUGGCACGGCGGAGUACGAUCAGAAAGCGAUCAGUUAGGUAAUUUA-AACC---AGCUUCAAUCUUAACCAUAGUCCGUAACGUGCCAAG-UA-UCAU---AACCAU------ .....((((((((((((((.((.((((.......))))((((.(((......-.)))---)))).............)).)))))..)))))).))-).-....---......------ ( -28.20) >DroMoj_CAF1 9947 105 - 1 GCAACACUAGCACGUCGGAGUACGAUCAGAAAGCGAUCAGUUAGGUAAUUUAAAAACA--AACAACA-CCUUAACUUUAACAUGUCAAGUGCAUAG-UA-UU------GCUAACGU--- ((((.((((((((((((.....))))..((........((((((((............--......)-)).)))))........))..)))).)))-).-))------))......--- ( -22.16) >DroAna_CAF1 7444 108 - 1 GCAACACUAGCACGGCGGAGUACGAUCAGAAAGCGAUCCGUUAGGUAAUUUA-AA-C---UGCUUCA-UCUAAACUAUAUUCCGUAAAGUGCCAAG-UA-CUAU---UGCCAAAGUACU .........((((.((((((((.((((.......))))..((((((......-..-.---......)-)))))....))))))))...))))..((-((-((..---......)))))) ( -25.36) >consensus GCAACACUAGCACGGCGGAGUACGAUCAGAAAGCGAUCAGUUAGGUAAUUUA_AACC___AGCUUCA_UCUUAACCAUAGUCCGUAAAGUGCAAAG_UA_UCAU___AACCAU______ .........((((.(((((.((.((((.......)))).((((((........................))))))..)).)))))...))))........................... (-15.81 = -16.48 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:18 2006