
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,759,304 – 11,759,461 |
| Length | 157 |
| Max. P | 0.972346 |

| Location | 11,759,304 – 11,759,401 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 87.41 |
| Mean single sequence MFE | -21.07 |
| Consensus MFE | -16.72 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11759304 97 - 22224390 ACAAAUUGGGAUAGCCGUUCUAAAUGAAUUGCAUAUAUUGAAUAUAUAUGUAUAUGCAUACAUUUGUCUUAGAUCAUAUGUAAUUUAAGGUUGGUUA ...........((((((..((...(((((((((((((((.((.(((.((((((....)))))).))).)).))).))))))))))))))..)))))) ( -21.80) >DroSec_CAF1 12709 92 - 1 ACAAAUUG-GACAGACGUUCCAAAUGAACUACAUAUAUUGAAUGUAUAUGCAUAU----ACAUAUGUCUCAGAUCUUAUAUAACUUAAGGUUUGUUA .......(-((((...((((.....))))..(((((((.....))))))).....----.....)))))(((((((((.......)))))))))... ( -18.30) >DroSim_CAF1 7620 92 - 1 ACAAAUUG-GACAGACGUUCCAAAUGAACUGCAUAUAUUGAAUGUAUAUGCAUAU----ACAUAUGUCUCAGAUCUUAUAUAACUUAAGGUUUGUUA .......(-((((...((((.....))))(((((((((.....)))))))))...----.....)))))(((((((((.......)))))))))... ( -23.10) >consensus ACAAAUUG_GACAGACGUUCCAAAUGAACUGCAUAUAUUGAAUGUAUAUGCAUAU____ACAUAUGUCUCAGAUCUUAUAUAACUUAAGGUUUGUUA .........((((...((((.....))))(((((((((.....)))))))))............)))).(((((((((.......)))))))))... (-16.72 = -16.73 + 0.01)

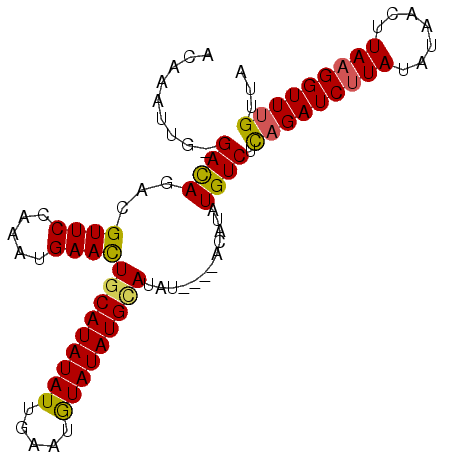

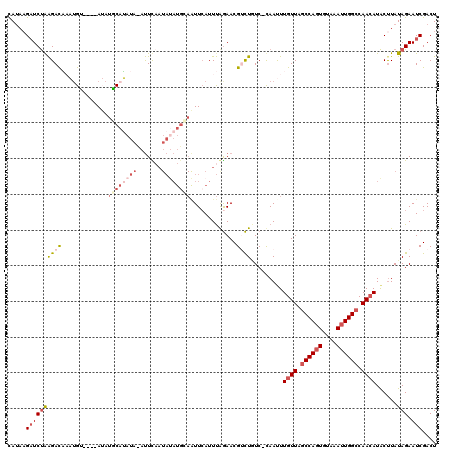
| Location | 11,759,321 – 11,759,441 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.94 |
| Mean single sequence MFE | -24.06 |
| Consensus MFE | -9.75 |
| Energy contribution | -12.65 |
| Covariance contribution | 2.90 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11759321 120 + 22224390 CAUAUGAUCUAAGACAAAUGUAUGCAUAUACAUAUAUAUUCAAUAUAUGCAAUUCAUUUAGAACGGCUAUCCCAAUUUGUUAGCCAGUGUAAAUUGGCCAAAAUACUUAUAGAAUCGACU ....(((((((.(((((((...((((((((.............)))))))).............((.....)).))))))).((((((....))))))...........))).))))... ( -22.72) >DroSec_CAF1 12726 115 + 1 UAUAAGAUCUGAGACAUAUGU----AUAUGCAUAUACAUUCAAUAUAUGUAGUUCAUUUGGAACGUCUGUC-CAAUUUGUUAGCCAGUGUAAAUUGGCCAACAUACUUAUAGAAUCGACU ((((((.....((((......----...((((((((.......))))))))((((.....))))))))...-.....((((.((((((....)))))).))))..))))))......... ( -28.10) >DroSim_CAF1 7637 115 + 1 UAUAAGAUCUGAGACAUAUGU----AUAUGCAUAUACAUUCAAUAUAUGCAGUUCAUUUGGAACGUCUGUC-CAAUUUGUUAGCCAGUGUAAAUUGGCCAACAUACUUAUAGAAUCGACU ((((((.....((((......----...((((((((.......))))))))((((.....))))))))...-.....((((.((((((....)))))).))))..))))))......... ( -30.10) >DroEre_CAF1 41881 99 + 1 CAUAAGAUCUAGGACCAUUGC----A---GCCGCC---CUU-----------UUCAUUCAAAACGUUUUUCGUAACUUGUUGGCCAUUGUAAAUUGGCCAACAUACUUAUAGAAUCGACU .....((...(((......((----.---...)))---)).-----------.))........((.((((.((((..(((((((((........)))))))))...)))))))).))... ( -20.80) >DroYak_CAF1 13913 108 + 1 CAAAAGCUCCAAGAUUA--GC----A---GCAGAU---UUUCAUACAUGCAAUUAAUUUAGAACGACUUUCAUAACUUGUUGACCAGUGUAAAUUGGCCAACAUGCCUAUAGAAUCGACU .....(((........)--))----(---(..(((---(((.(((...(((.........(((.....)))......(((((.(((((....))))).)))))))).)))))))))..)) ( -18.60) >consensus CAUAAGAUCUAAGACAAAUGU____AUAUGCAUAUA_AUUCAAUAUAUGCAAUUCAUUUAGAACGUCUGUC_CAAUUUGUUAGCCAGUGUAAAUUGGCCAACAUACUUAUAGAAUCGACU .....((((((((((.............((((((((.......)))))))).............)))).........((((.((((((....)))))).))))......))).))).... ( -9.75 = -12.65 + 2.90)

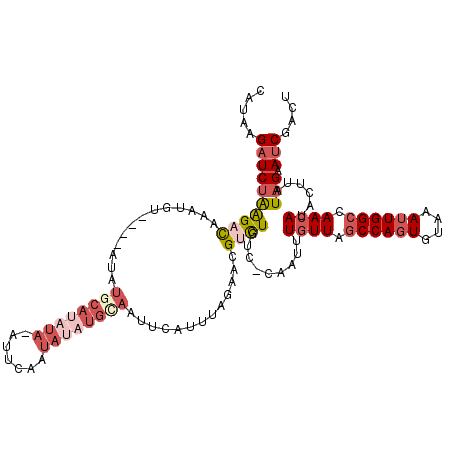
| Location | 11,759,321 – 11,759,441 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.94 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -15.41 |
| Energy contribution | -17.52 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11759321 120 - 22224390 AGUCGAUUCUAUAAGUAUUUUGGCCAAUUUACACUGGCUAACAAAUUGGGAUAGCCGUUCUAAAUGAAUUGCAUAUAUUGAAUAUAUAUGUAUAUGCAUACAUUUGUCUUAGAUCAUAUG ....(((.(((.........((((((........))))))((((((((((((....)))))))(((...(((((((((.....)))))))))....)))...)))))..))))))..... ( -24.60) >DroSec_CAF1 12726 115 - 1 AGUCGAUUCUAUAAGUAUGUUGGCCAAUUUACACUGGCUAACAAAUUG-GACAGACGUUCCAAAUGAACUACAUAUAUUGAAUGUAUAUGCAUAU----ACAUAUGUCUCAGAUCUUAUA .........((((((..(((((((((........)))))))))..(((-((((...((((.....))))..(((((((.....))))))).....----.....)))).)))..)))))) ( -27.10) >DroSim_CAF1 7637 115 - 1 AGUCGAUUCUAUAAGUAUGUUGGCCAAUUUACACUGGCUAACAAAUUG-GACAGACGUUCCAAAUGAACUGCAUAUAUUGAAUGUAUAUGCAUAU----ACAUAUGUCUCAGAUCUUAUA .........((((((..(((((((((........)))))))))..(((-((((...((((.....))))(((((((((.....)))))))))...----.....)))).)))..)))))) ( -31.90) >DroEre_CAF1 41881 99 - 1 AGUCGAUUCUAUAAGUAUGUUGGCCAAUUUACAAUGGCCAACAAGUUACGAAAAACGUUUUGAAUGAA-----------AAG---GGCGGC---U----GCAAUGGUCCUAGAUCUUAUG (((((.((((.......(((((((((........))))))))).....(((((....)))))......-----------.))---))))))---)----.....((((...))))..... ( -23.70) >DroYak_CAF1 13913 108 - 1 AGUCGAUUCUAUAGGCAUGUUGGCCAAUUUACACUGGUCAACAAGUUAUGAAAGUCGUUCUAAAUUAAUUGCAUGUAUGAAA---AUCUGC---U----GC--UAAUCUUGGAGCUUUUG ....((((.....(((.(((((((((........))))))))).))).....))))(((((((((((...(((.(.((....---))))))---.----..--)))).)))))))..... ( -26.20) >consensus AGUCGAUUCUAUAAGUAUGUUGGCCAAUUUACACUGGCUAACAAAUUG_GAAAGACGUUCUAAAUGAACUGCAUAUAUUGAAU_UAUAUGCAUAU____ACAUUUGUCUUAGAUCUUAUG .........((((((..(((((((((........)))))))))..(((.((((...((((.....))))(((((((((.....)))))))))............)))).)))..)))))) (-15.41 = -17.52 + 2.11)



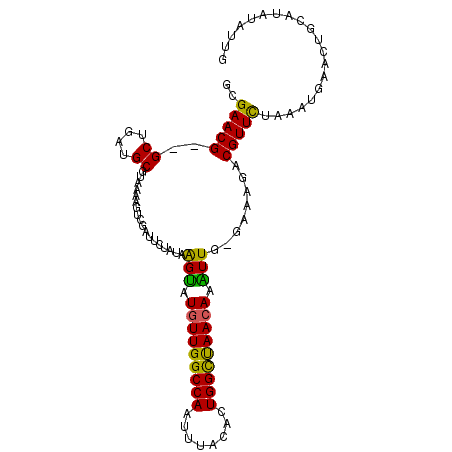
| Location | 11,759,361 – 11,759,461 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 84.30 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -19.48 |
| Energy contribution | -18.56 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11759361 100 - 22224390 GCGAACG--GCUGAUGCGUAAAAGUCGAUUCUAUAAGUAUUUUGGCCAAUUUACACUGGCUAACAAAUUGGGAUAGCCGUUCUAAAUGAAUUGCAUAUAUUG ..(((((--((((((........))).((((((........(((((((........))))))).....)))))))))))))).................... ( -25.02) >DroSec_CAF1 12762 101 - 1 GCGAACGGGGCUGAUGCGUAAAAGUCGAUUCUAUAAGUAUGUUGGCCAAUUUACACUGGCUAACAAAUUG-GACAGACGUUCCAAAUGAACUACAUAUAUUG (.((((...((....))......(((...((((...((.(((((((((........))))))))).))))-))..))))))))................... ( -23.20) >DroSim_CAF1 7673 101 - 1 GCGAACGGGGCUGAUGCGUAAAAGUCGAUUCUAUAAGUAUGUUGGCCAAUUUACACUGGCUAACAAAUUG-GACAGACGUUCCAAAUGAACUGCAUAUAUUG ((.......))..((((......(((...((((...((.(((((((((........))))))))).))))-))..)))((((.....)))).))))...... ( -24.40) >DroEre_CAF1 41911 89 - 1 GCGAACG--GCUGAUGCGUAAAAGUCGAUUCUAUAAGUAUGUUGGCCAAUUUACAAUGGCCAACAAGUUACGAAAAACGUUUUGAAUGAA-----------A ..(((((--(((..........))))).)))........(((((((((........))))))))).....(((((....)))))......-----------. ( -22.10) >DroYak_CAF1 13941 100 - 1 GCGAACG--GCUGAUGCGUAAAAGUCGAUUCUAUAGGCAUGUUGGCCAAUUUACACUGGUCAACAAGUUAUGAAAGUCGUUCUAAAUUAAUUGCAUGUAUGA ..(((((--(((....(((((..(((.........))).(((((((((........)))))))))..)))))..)))))))).................... ( -28.80) >consensus GCGAACG__GCUGAUGCGUAAAAGUCGAUUCUAUAAGUAUGUUGGCCAAUUUACACUGGCUAACAAAUUG_GAAAGACGUUCUAAAUGAACUGCAUAUAUUG ..(((((..((....))..................(((.(((((((((........))))))))).)))........))))).................... (-19.48 = -18.56 + -0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:06 2006