| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,310,657 – 1,310,757 |
| Length | 100 |
| Max. P | 0.653857 |

| Location | 1,310,657 – 1,310,757 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 76.63 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.45 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
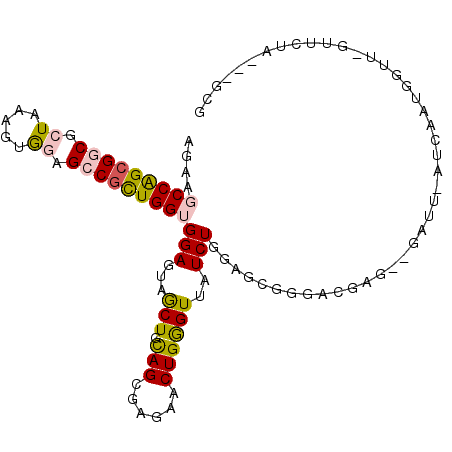
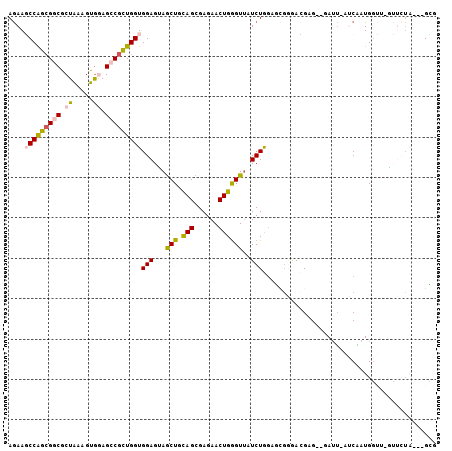
>X_DroMel_CAF1 1310657 100 - 22224390 AGAAGCCAGCGGCGCUGAAGUGGAGCCGCUGGUGGAGUAGCUGCAGCGAGAACUGGGUUAUCUGGAGCGGGACGAG--GAUU-AUCAAUGGCGCACAAUA---GCG .((.(((((((((.((.....)).)))))))))...((.(((.(((....(((...)))..))).)))...))...--....-.)).....(((......---))) ( -31.10) >DroSec_CAF1 46973 99 - 1 AGAAGCCAGCGGCGCUAAAGUGGAGCCGCUGGUGGAGUAGCUGCAGCGAGAACUGGGUUAUCUGGAGCGGGACGUG--GAUU-AUCAAUGGUU-GUUCUA---GCG ....(((((((((.((.....)).)))))))))...((((((.(((......)))))))))(((((((((..(((.--(...-..).))).))-))))))---).. ( -35.20) >DroSim_CAF1 35108 99 - 1 AGAAGCCAGCGGCGCUAAAGUGGAGCCGCUGGUGGAGGAGCUGCAGCGAGAACUGGGUUAUCUGGAGCGGGACGUG--GAUU-AUCAAUGGUU-GUUCUA---GCG ....(((((((((.((.....)).))))))))).....((((.(((......)))))))..(((((((((..(((.--(...-..).))).))-))))))---).. ( -33.20) >DroEre_CAF1 44955 99 - 1 AGAAGCCAGCGGCGCUGAAGUGGAGCCGUUGGUGGAGUAGCUGCAGCGAGAACUGGGUUAUCUGGCGAGAGACGGG--UAUU-AUCAAUGGUU-GCUUUG---GUG ....(((((((((.((.....)).)))))))))(((((((((.(((......)))(((((((((.(....).))))--))..-)))...))))-))))).---... ( -34.80) >DroAna_CAF1 40968 102 - 1 AGAAUCCGGAGGCACUAAAGUGAAGGCGCUGGUGGAGCAACUGCAGCGAGAACUGAGUUAUCUAGACCGGAAAGAGAGAAUUUUACGAUGUGG-GAUCUA---GGA ....(((..(((..(((..((((((.(.((((((((..((((.(((......))))))).)))..))))).....)....))))))....)))-..))).---))) ( -23.30) >DroPer_CAF1 64389 105 - 1 AAAAUCCGGCGGCGCUGAAAUAUAGGCGCUGGUGGAGGAGCUGUAGCGAGAACUGGGUGAUCUAACGAUCAAGGGGAGAACAGAGCUAUGGUA-UUACAAGAAGCA ....((((.(((((((........))))))).))))...((((((((.....((...(((((....))))).))(.....)...)))))))).-............ ( -35.30) >consensus AGAAGCCAGCGGCGCUAAAGUGGAGCCGCUGGUGGAGUAGCUGCAGCGAGAACUGGGUUAUCUGGAGCGGGACGAG__GAUU_AUCAAUGGUU_GUUCUA___GCG ....(((((((((.((.....)).)))))))))(((...(((.(((......))))))..)))........................................... (-18.20 = -18.45 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:44 2006