
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,681,509 – 11,681,646 |
| Length | 137 |
| Max. P | 0.998065 |

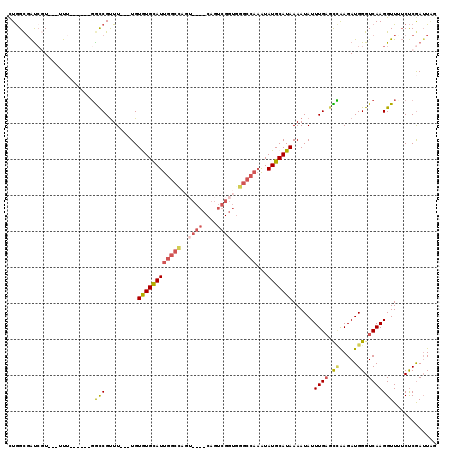
| Location | 11,681,509 – 11,681,616 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.30 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -15.91 |
| Energy contribution | -16.92 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11681509 107 - 22224390 UUGGCGAUCGU---UUU------GGUCGUUU---UGUGUGUAUUGGCCAGUUGGCCAGUCGGUGGGCCAAAUAUGCAUAAAAUAUUUGAGCCAAGAUGGGUCAAGGUUUUCUCGAUUAG (((((..((((---(((------((((((((---((((((((((((((..((((....))))..)))).))))))))))))))....)).))))))))))))))............... ( -35.90) >DroPse_CAF1 7721 107 - 1 CUAGCCAUCAUCGCUUAACACAUCAUCGCUUAAAUGUAUGCAUUGGCUUGC------------CGGCCAAGUAUGCAUAAAAUAUUUGACUGGAGAUGGGUCAAGGUUGUCCCAAAUUG ....((((((..((.............))....((((((((.((((((...------------.))))))))))))))........))).)))...((((.((....)).))))..... ( -26.92) >DroSec_CAF1 7996 107 - 1 UUGGCGAUCGU---UUG------GGCCGUUU---UGUGUGCAUUGGCCAGUUGGCCAGUCGGUGGGCCAAAUAUGCAUAAAAUAUUUGAGCCAAGAUGGGUCAAGGUUUUCUCGAUUAG .....(((((.---..(------((((((((---((((((.(((((((....))))))))((....))......)))))))))..((((.((.....)).)))))))))...))))).. ( -36.80) >DroSim_CAF1 9846 107 - 1 UUGGCGAUCGU---UUU------GGCCGUUU---UGUGUGCAUUGGCCAGUUGGCCAGUCGGUGGGCCAAAUAUGCAUAAAAUAUUUGAGCCAAGAUGGGUCAAGGUUUUCUCGAUUAG .....(((((.---...------((((((((---((((((.(((((((....))))))))((....))......)))))))))..((((.((.....)).))))))))....))))).. ( -36.00) >DroEre_CAF1 7353 100 - 1 CUGGUGGC--U---UUU------GGCCGUUU---UGUGUGCAUUGGCUAGU----CAGCCGGUGGGCCAAAUAUGCAUAAAAUAUUUGUGUUAAGAUGGGUCAAGGUU-GCUCGAUUGU (.((..((--(---((.------(.((((((---((.((.((((((((...----.)))))))).))(((((((.......)))))))...)))))))).).))))).-.)).)..... ( -34.00) >DroYak_CAF1 10623 87 - 1 CUGGCGAUCGU---UUU------GGCCGUUU---UGUAUGCAUUGGCUAGC-------------------GUAUGCAUAAAAUAUUUGAGUCAAGAUGGGUCAAGGUU-UCUCGAUUAU .((((..((((---(((------(((((..(---..(((((((..(....)-------------------..)))))))....)..)).)))))))))))))).....-.......... ( -20.10) >consensus CUGGCGAUCGU___UUU______GGCCGUUU___UGUGUGCAUUGGCCAGU____CAGUCGGUGGGCCAAAUAUGCAUAAAAUAUUUGAGCCAAGAUGGGUCAAGGUUUUCUCGAUUAG ........................(((.........((((((((((((..((((....))))..)))))...)))))))......((((.((.....)).)))))))............ (-15.91 = -16.92 + 1.00)


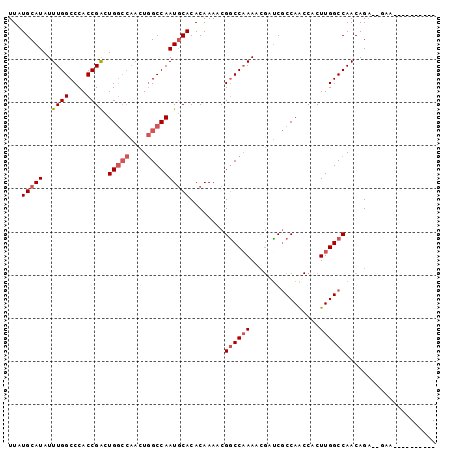
| Location | 11,681,548 – 11,681,646 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -23.45 |
| Consensus MFE | -15.53 |
| Energy contribution | -16.84 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11681548 98 + 22224390 UUAUGCAUAUUUGGCCCACCGACUGGCCAACUGGCCAAUACACACAAAACGACCAAAACGAUCGCCAACCACUUGGCCAACAGAC-GAAAAUUCAGACG ..........((((((.......(((((....)))))............(((.(.....).)))..........)))))).....-............. ( -18.60) >DroSec_CAF1 8035 88 + 1 UUAUGCAUAUUUGGCCCACCGACUGGCCAACUGGCCAAUGCACACAAAACGGCCCAAACGAUCGCCAACCACUUGGCCAACAGAC-GCA---------- ...(((....((((((........)))))).(((((((((....))....(((..........)))......)))))))......-)))---------- ( -23.50) >DroSim_CAF1 9885 83 + 1 UUAUGCAUAUUUGGCCCACCGACUGGCCAACUGGCCAAUGCACACAAAACGGCCAAAACGAUCGCCAACCACUUGGCCAACAG---------------- ...(((((..((((....)))).(((((....))))))))))........((((((................)))))).....---------------- ( -23.79) >DroEre_CAF1 7391 91 + 1 UUAUGCAUAUUUGGCCCACCGGCUG----ACUAGCCAAUGCACACAAAACGGCCAAAA--GCCACCAG-CACUUGGCCAACAGA-CAAAAGGCCGAAAG .........(((((((....((((.----...))))..............((((((..--((.....)-)..))))))......-.....))))))).. ( -27.90) >consensus UUAUGCAUAUUUGGCCCACCGACUGGCCAACUGGCCAAUGCACACAAAACGGCCAAAACGAUCGCCAACCACUUGGCCAACAGA__GAA__________ ...(((((..((((....)))).(((((....))))))))))........((((((................))))))..................... (-15.53 = -16.84 + 1.31)


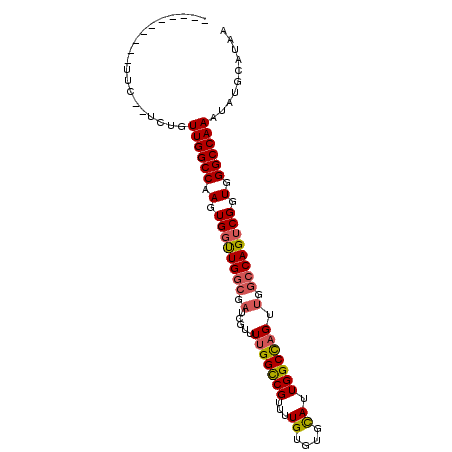
| Location | 11,681,548 – 11,681,646 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -34.35 |
| Consensus MFE | -25.50 |
| Energy contribution | -26.00 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.74 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11681548 98 - 22224390 CGUCUGAAUUUUC-GUCUGUUGGCCAAGUGGUUGGCGAUCGUUUUGGUCGUUUUGUGUGUAUUGGCCAGUUGGCCAGUCGGUGGGCCAAAUAUGCAUAA ((.(..((....(-(..(((..(((....)))..)))..)).))..).))..(((((((((((((((..((((....))))..)))).))))))))))) ( -34.90) >DroSec_CAF1 8035 88 - 1 ----------UGC-GUCUGUUGGCCAAGUGGUUGGCGAUCGUUUGGGCCGUUUUGUGUGCAUUGGCCAGUUGGCCAGUCGGUGGGCCAAAUAUGCAUAA ----------(((-((...((((((.(.((..((((.(.(.....(((((...((....)).))))).).).))))..)).).))))))..)))))... ( -36.00) >DroSim_CAF1 9885 83 - 1 ----------------CUGUUGGCCAAGUGGUUGGCGAUCGUUUUGGCCGUUUUGUGUGCAUUGGCCAGUUGGCCAGUCGGUGGGCCAAAUAUGCAUAA ----------------...((((((.(.((..((((.(.....(((((((...((....)).))))))).).))))..)).).)))))).......... ( -32.20) >DroEre_CAF1 7391 91 - 1 CUUUCGGCCUUUUG-UCUGUUGGCCAAGUG-CUGGUGGC--UUUUGGCCGUUUUGUGUGCAUUGGCUAGU----CAGCCGGUGGGCCAAAUAUGCAUAA .....(((((.(((-.((((((((((((..-(.((((((--.....))))))....)..).)))))))).----))).))).)))))............ ( -34.30) >consensus __________UUC__UCUGUUGGCCAAGUGGUUGGCGAUCGUUUUGGCCGUUUUGUGUGCAUUGGCCAGUUGGCCAGUCGGUGGGCCAAAUAUGCAUAA ...................((((((.(.((((((((.(.....(((((((...((....)).))))))).).)))))))).).)))))).......... (-25.50 = -26.00 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:46 2006