| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,303,201 – 1,303,300 |
| Length | 99 |
| Max. P | 0.845518 |

| Location | 1,303,201 – 1,303,300 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -17.63 |
| Energy contribution | -17.43 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
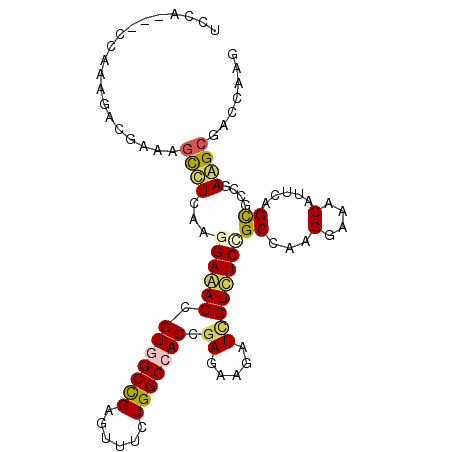
>X_DroMel_CAF1 1303201 99 + 22224390 GCCA---CCAAAGACGAUAGCUUAAAGGAAACUGUGGCCGAGUUUCUGGCCACCGAGAAGAUUGUUUCCGCCAAGGAAGCCUUCAGCACUGAAGCCACUAAG ....---............((((..((....))(((((((......)))))))...((((...((((((.....)))))))))).......))))....... ( -27.40) >DroPse_CAF1 56171 99 + 1 UGCA---CCAGAGAUGAAAGUCUCAAGGAGACCGUGGCCGAGUUUCUGGCCACGGAGAAGAUCGUCUCGGCCAAGGAAACAUUCAGUGCCGAGGCUUCCAAG ....---...(((((....)))))..((((.(((((((((......)))))))))........(((((((((..(....).....).))))))))))))... ( -44.20) >DroEre_CAF1 37342 99 + 1 GCCA---CCAAAGACGACAGCUUAAAGGAAACUGUGGCCGAGUUCCUGGCCACCGAGAAGAUUGUCUCCGCCAAGGAAGCCUUCAGCGCUGAAGCGACCAAG ....---.....((.((((((((..((....))(((((((......)))))))....))).)))))))......((..((.((((....))))))..))... ( -28.80) >DroMoj_CAF1 54007 97 + 1 AUCAG--UAA---AGGACAGUCUCAAGGAAACGGUAGCUGAAUUUUUGGCAACGGAGAAAAUAGUUUCUGCCAAGGAAACCUUCAGCGCUGAGGCAACCAAA .....--...---.((...((((((.(....)....((((((((((((....)))))).....((((((.....)))))).))))))..))))))..))... ( -29.10) >DroAna_CAF1 32368 102 + 1 UCCAGUUCCAAGGAGGAGAACCUGAAGGAGACAGUCGCCGAGUUCCUGGCCACCGAGAAGAUCGUCUCCGCCAAGGAGACAUUCAGCGCCGAAGCGGCCAAA (((........)))((((....(((.(....)..))).....))))(((((........((..((((((.....))))))..)).((......))))))).. ( -31.60) >DroPer_CAF1 55861 99 + 1 UGCA---CCAGAGAUGAAAGUCUCAAGGAGACCGUGGCCGAGUUUCUGGCCACGGAGAAGAUCGUCUCGGCCAAGGAAACAUUCAGUGCCGAGGCUUCCAAG ....---...(((((....)))))..((((.(((((((((......)))))))))........(((((((((..(....).....).))))))))))))... ( -44.20) >consensus UCCA___CCAAAGACGAAAGCCUCAAGGAAACCGUGGCCGAGUUUCUGGCCACCGAGAAGAUCGUCUCCGCCAAGGAAACAUUCAGCGCCGAAGCGACCAAG ...................((((...((((((.(((((((......))))))).((.....))))))))((...(....).....))....))))....... (-17.63 = -17.43 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:42 2006