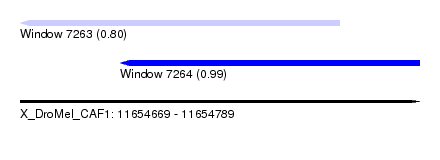
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,654,669 – 11,654,789 |
| Length | 120 |
| Max. P | 0.988759 |

| Location | 11,654,669 – 11,654,765 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 81.49 |
| Mean single sequence MFE | -21.58 |
| Consensus MFE | -14.90 |
| Energy contribution | -15.86 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11654669 96 - 22224390 AGCGAGCGAAAGAGAGACAAUCGACCACCCCCUCGCUGUUUCUCGGUCGCUUCCACACACACACACACGGCGACCAUAAGUAACGCAUUUGUCUUU .....(((...((((((((..(((........))).))))))))(((((((.................)))))))........))).......... ( -26.93) >DroSec_CAF1 27915 94 - 1 AGCGAGCAAAAGAGAGACAAUCGACCACCCCCUCGCUGUUUCUCGCUCGCUUCCACACAC--ACACACGACGACCAUAAGUAACGCAUUUGUCUUU (((((((....((((((((..(((........))).))))))))))))))).........--......(((((...............)))))... ( -23.36) >DroSim_CAF1 24414 92 - 1 AGCGAGCGAAAGAGAGACCAUCGACCACCCCCUCCCUGUUUCUCGCUCGCUUCCACACA----CACACGACGACCAUACGUAACGCCUUUGUCUUU .((((((((..(((((((....((........))...)))))))..)))))........----......(((......)))..))).......... ( -19.40) >DroEre_CAF1 30107 72 - 1 UGCGAGAGAAAGAGAGACAAUCGACCACCCCCUCGCUGUUUCGCGCUCGCUUCCACAC------------------------ACGCAUUUGUCUUU .(((((.(.....((((((..(((........))).)))))).).)))))........------------------------.............. ( -14.80) >DroYak_CAF1 27380 90 - 1 UGCGAGAGAAAUAGAGACAAUCGACCACCCCCUCGCUGUUUCUCUCUCGCUCGCACUC------ACACAGCGACCAUAAGUAACGCAUUUGCCUUU .((((((((....((((((..(((........))).))))))))))))))((((....------.....))))...........((....)).... ( -23.40) >consensus AGCGAGCGAAAGAGAGACAAUCGACCACCCCCUCGCUGUUUCUCGCUCGCUUCCACACA____CACACGACGACCAUAAGUAACGCAUUUGUCUUU .((((((....((((((((..(((........))).)))))))))))))).............................................. (-14.90 = -15.86 + 0.96)



| Location | 11,654,699 – 11,654,789 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 87.56 |
| Mean single sequence MFE | -20.30 |
| Consensus MFE | -14.90 |
| Energy contribution | -15.86 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11654699 90 - 22224390 ACACAAACCUAAAAGCGACAU-ACUAGCGAGCGAAAGAGAGACAAUCGACCACCCCCUCGCUGUUUCUCGGUCGCUUCCACACACACACAC ..............((.....-....))((((((..((((((((..(((........))).))))))))..)))))).............. ( -22.30) >DroSec_CAF1 27945 88 - 1 ACACAAACCUAAAAGCGACAU-ACUAGCGAGCAAAAGAGAGACAAUCGACCACCCCCUCGCUGUUUCUCGCUCGCUUCCACACAC--ACAC .....................-...(((((((....((((((((..(((........))).))))))))))))))).........--.... ( -21.50) >DroSim_CAF1 24444 87 - 1 ACACAAACCUAAAAGCGACAUUACUAGCGAGCGAAAGAGAGACCAUCGACCACCCCCUCCCUGUUUCUCGCUCGCUUCCACACA----CAC .........................(((((((((.(((.((...................)).))).)))))))))........----... ( -17.41) >DroEre_CAF1 30121 82 - 1 ACACAAACUUAAAAGCGACAU-GCUUGCGAGAGAAAGAGAGACAAUCGACCACCCCCUCGCUGUUUCGCGCUCGCUUCCACAC-------- ............((((.....-))))(((((.(.....((((((..(((........))).)))))).).)))))........-------- ( -17.20) >DroYak_CAF1 27410 84 - 1 ACACAAACUCAAAAGCGACAU-ACUUGCGAGAGAAAUAGAGACAAUCGACCACCCCCUCGCUGUUUCUCUCUCGCUCGCACUC------AC ..............((((...-....((((((((....((((((..(((........))).))))))))))))))))))....------.. ( -23.11) >consensus ACACAAACCUAAAAGCGACAU_ACUAGCGAGCGAAAGAGAGACAAUCGACCACCCCCUCGCUGUUUCUCGCUCGCUUCCACACA____CAC ..........................((((((....((((((((..(((........))).))))))))))))))................ (-14.90 = -15.86 + 0.96)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:37 2006