
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,652,667 – 11,652,777 |
| Length | 110 |
| Max. P | 0.593378 |

| Location | 11,652,667 – 11,652,777 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -20.81 |
| Energy contribution | -21.59 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11652667 110 + 22224390 AAUAAAGAG-------CUUAGCUGCAAAAGAGAGGACAGC-AACAAAAUUUCGUUGCAGCUGAGUUGCUGAAAAGUGCGUUUGUGUGUGCAUUCAAGUGCAACUGGCUGUGUG--UUAGA .(((((.((-------(((((((((((..((((.......-.......)))).)))))))))))))(((....)))...)))))...(((((....))))).(((((.....)--)))). ( -31.74) >DroSec_CAF1 25989 109 + 1 AAUAAAGAU-------CUUAGCUGCAAAAGAGAGGACAGC-AACAAAAUUUCGUUGCAGCUGAGUUGCUGAAAAAUGCGUUUGUGUGUGCAUUCAAGUGCAACUGGCUGUG-G--UUAGA .........-------.(((((..(.............((-(((........)))))((((.((((((.....((((((........)))))).....)))))))))))..-)--)))). ( -30.50) >DroSim_CAF1 22073 110 + 1 AAUAAAGAG-------CUUAGCUGCAAAAGAGAGGACAGC-AACAAAAUUUCGUUGCAGCUGAGUUGCUGAAAAAUGCGUUUGUGUGUGCAUUCAAGUGCAACUGGCUGUGUG--UUAGA ((((.(.((-------(((((((((((..((((.......-.......)))).)))))))))((((((.....((((((........)))))).....)))))))))).).))--))... ( -31.14) >DroEre_CAF1 28158 106 + 1 ----AAGAG-------CUUAGCUGCAAAAGAGAGGAUAGC-AACAACAUUUCGUUGCAGCUGAGUUGCUGAAAAGUGCGUUUGUGUGUGCAUUCAAGUGCAACUGGCUGUGUG--UUAGA ----...((-------(((((((((((..((((.......-.......)))).)))))))))))))(((....)))(((((((.((....)).)))))))..(((((.....)--)))). ( -31.34) >DroYak_CAF1 25439 109 + 1 AAU-AAGAG-------CAUAGCUGCAAAAGAGAGGACACC-AACAACAUUUCGUUGCAGCUGAGUUGCUGAAAAGUGCGUUUGUGUGUGCAUUCAAGUGCAACUGGCAGUGUG--UUAGA ...-...((-------((((.((((...((.....((((.-(((.((.(((((..(((((...)))))))))).))..))).)))).(((((....))))).)).))))))))--))... ( -30.60) >DroPer_CAF1 27527 107 + 1 AGCUAGGUGGAUUUGUUUUGGGU--GGGGGAGAGUGAAGCAAACAACAUUUCGUUGCAGCUGAGUGAAUGGC-AUUGCGCUUGUGUGUGCAU----------CUGCCUGUGUGCGUUAAG .((((((..((((..(((...(.--.(.((((.((..........)).)))).)..)....)))..))....-..(((((......))))))----------)..))))...))...... ( -25.30) >consensus AAUAAAGAG_______CUUAGCUGCAAAAGAGAGGACAGC_AACAAAAUUUCGUUGCAGCUGAGUUGCUGAAAAGUGCGUUUGUGUGUGCAUUCAAGUGCAACUGGCUGUGUG__UUAGA ..................(((((((((..((((...............)))).)))))))))((((((.....(((((((......))))))).....))))))................ (-20.81 = -21.59 + 0.78)

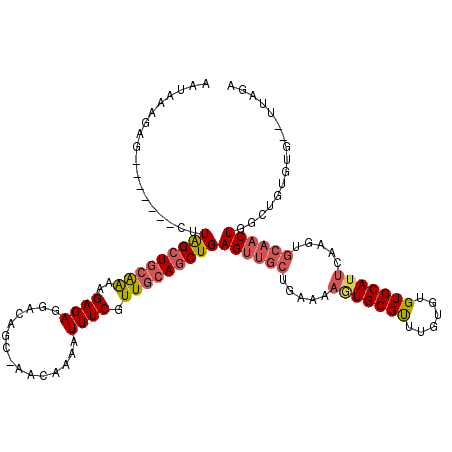
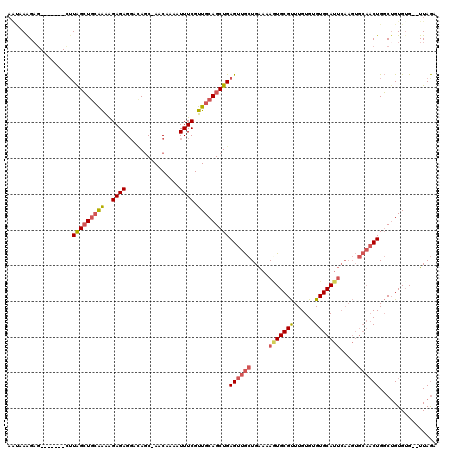
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:30 2006