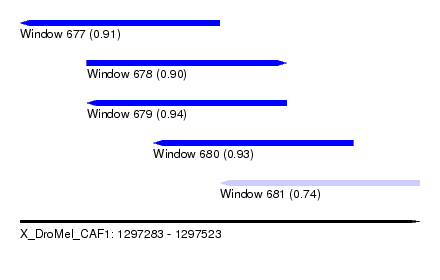
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,297,283 – 1,297,523 |
| Length | 240 |
| Max. P | 0.936514 |

| Location | 1,297,283 – 1,297,403 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -44.45 |
| Consensus MFE | -41.88 |
| Energy contribution | -41.25 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1297283 120 - 22224390 UAUGCUCUGGAGCAGAUGGAGUUCUUAGAGCCUCGUCUAAAGGAGACGAGACCGUUUUAGCUGCACCAAUCUGUGCGAGUUCAGCAGGUUUCGUAAGUGUGGGCGUGACGGUGGAAACCG ...((((((((((.......)))).))))))(.((((((......(((((((((((..((((((((......)))).)))).))).)))))))).....)))))).).((((....)))) ( -44.80) >DroSec_CAF1 34288 120 - 1 UAUGCUCUGUAGCAGAUGGAGUUCUUAGAGCCUCGUCUACAGGAGACGAGACCGUUAUAGCUGCACCAAUCUUUGCGAGUUCUGCAGGUUUCGUAAGUGUGGGCGUGACGGUGGAAACCG (((((..((((((.(((((.((((...))))(((((((.....))))))).)))))...))))))(((...((((((((..(....)..))))))))..)))))))).((((....)))) ( -43.80) >DroSim_CAF1 23151 120 - 1 UAUGCUCUGGAGCAGAUGGAGUUCUUGGAGCCUCGUCUACAGGAGACGAGACCGUUAUAGCUGCACCAAUCUUUGCGAGUUCUGCAGGUUUCGUAAGUGUGGGCGUGACGGUGGAAACCG ...((((((((((.......)))).))))))(.((((((((....((((((((((...(((((((........))).))))..)).))))))))...)))))))).).((((....)))) ( -44.20) >DroYak_CAF1 30836 120 - 1 UAUGCUCUGGAGCAGAUGGAGUUUUUGGAGCCUCGUCCACAGGAGACGAGACCGUUAUAGCUGCGCCAAUCUUUGCGAGUUCCGCAGGUUUUGUAAGUGUGGGCGGGACGGUGGAAACUG ...((((..((((.......).)))..))))((((((((((....((((((((.....((((.(((........))))))).....))))))))...)))))))))).((((....)))) ( -45.00) >consensus UAUGCUCUGGAGCAGAUGGAGUUCUUAGAGCCUCGUCUACAGGAGACGAGACCGUUAUAGCUGCACCAAUCUUUGCGAGUUCUGCAGGUUUCGUAAGUGUGGGCGUGACGGUGGAAACCG ...((((((((((.......)))).))))))(.((((((((....((((((((.....(((((((........))).)))).....))))))))...)))))))).).((((....)))) (-41.88 = -41.25 + -0.62)

| Location | 1,297,323 – 1,297,443 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -33.40 |
| Energy contribution | -34.02 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1297323 120 + 22224390 ACUCGCACAGAUUGGUGCAGCUAAAACGGUCUCGUCUCCUUUAGACGAGGCUCUAAGAACUCCAUCUGCUCCAGAGCAUAUAUCCAGAGCAGAUUCACCCGCAGAAUGUGCGAGUGAAGA (((((((((....((((..........((((((((((.....))))))))))...........((((((((..((.......))..)))))))).)))).......)))))))))..... ( -44.90) >DroSec_CAF1 34328 120 + 1 ACUCGCAAAGAUUGGUGCAGCUAUAACGGUCUCGUCUCCUGUAGACGAGGCUCUAAGAACUCCAUCUGCUACAGAGCAUAUAUCCCGAGCACAUUCACCCGCAGAAUAUGCGAGUGAAGA .((((....(((..((((.........((((((((((.....))))))))))............((((...))))))))..))).))))....(((((.((((.....)))).))))).. ( -36.30) >DroSim_CAF1 23191 120 + 1 ACUCGCAAAGAUUGGUGCAGCUAUAACGGUCUCGUCUCCUGUAGACGAGGCUCCAAGAACUCCAUCUGCUCCAGAGCAUAUAUCCCGAGCAGAUUCACCCGCAGAAUAUGCGAGUGAAGA (((((((......((((..........((((((((((.....))))))))))...........((((((((..((.......))..)))))))).)))).........)))))))..... ( -38.86) >DroYak_CAF1 30876 120 + 1 ACUCGCAAAGAUUGGCGCAGCUAUAACGGUCUCGUCUCCUGUGGACGAGGCUCCAAAAACUCCAUCUGCUCCAGAGCAUAUAUCCCGUGCAGAUUCACCCGCAGAAUGUGCGACUGAAGA ..(((((.......(((..........((((((((((.....))))))))))...........((((((.(..((.......))..).)))))).....)))......)))))....... ( -32.02) >consensus ACUCGCAAAGAUUGGUGCAGCUAUAACGGUCUCGUCUCCUGUAGACGAGGCUCCAAGAACUCCAUCUGCUCCAGAGCAUAUAUCCCGAGCAGAUUCACCCGCAGAAUAUGCGAGUGAAGA (((((((.......(((..........((((((((((.....))))))))))...........((((((((..((.......))..)))))))).....)))......)))))))..... (-33.40 = -34.02 + 0.62)

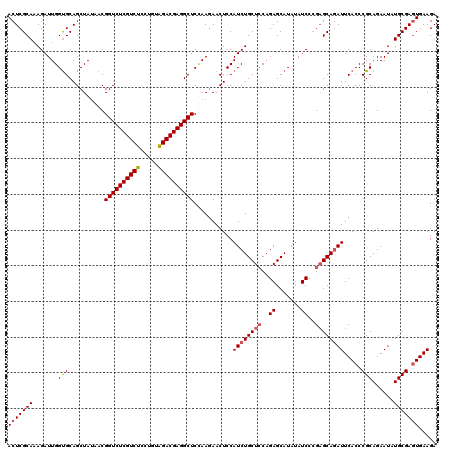
| Location | 1,297,323 – 1,297,443 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -44.25 |
| Consensus MFE | -38.03 |
| Energy contribution | -38.90 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1297323 120 - 22224390 UCUUCACUCGCACAUUCUGCGGGUGAAUCUGCUCUGGAUAUAUGCUCUGGAGCAGAUGGAGUUCUUAGAGCCUCGUCUAAAGGAGACGAGACCGUUUUAGCUGCACCAAUCUGUGCGAGU .....(((((((((...(((((.(((((((((((..((.......))..))))))))((.((((...))))(((((((.....))))))).))...))).)))))......))))))))) ( -51.50) >DroSec_CAF1 34328 120 - 1 UCUUCACUCGCAUAUUCUGCGGGUGAAUGUGCUCGGGAUAUAUGCUCUGUAGCAGAUGGAGUUCUUAGAGCCUCGUCUACAGGAGACGAGACCGUUAUAGCUGCACCAAUCUUUGCGAGU ..((((((((((.....))))))))))...(((((.((....((...((((((.(((((.((((...))))(((((((.....))))))).)))))...)))))).))....)).))))) ( -44.00) >DroSim_CAF1 23191 120 - 1 UCUUCACUCGCAUAUUCUGCGGGUGAAUCUGCUCGGGAUAUAUGCUCUGGAGCAGAUGGAGUUCUUGGAGCCUCGUCUACAGGAGACGAGACCGUUAUAGCUGCACCAAUCUUUGCGAGU ...(((((((((.....)))))))))((((((((.(((.......))).))))))))((((...((((.(((((((((.....)))))))...((....)).)).)))).))))...... ( -45.50) >DroYak_CAF1 30876 120 - 1 UCUUCAGUCGCACAUUCUGCGGGUGAAUCUGCACGGGAUAUAUGCUCUGGAGCAGAUGGAGUUUUUGGAGCCUCGUCCACAGGAGACGAGACCGUUAUAGCUGCGCCAAUCUUUGCGAGU ...(((.(((((.....))))).)))((((((.(.(((.......))).).))))))((((...((((.(((((((((....).))))))...((....)).)).)))).))))...... ( -36.00) >consensus UCUUCACUCGCACAUUCUGCGGGUGAAUCUGCUCGGGAUAUAUGCUCUGGAGCAGAUGGAGUUCUUAGAGCCUCGUCUACAGGAGACGAGACCGUUAUAGCUGCACCAAUCUUUGCGAGU ...(((((((((.....)))))))))((((((((.(((.......))).))))))))((.((((...))))(((((((.....))))))).))......((((((........))).))) (-38.03 = -38.90 + 0.87)

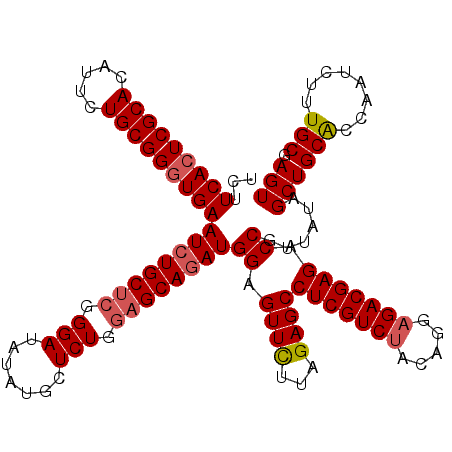
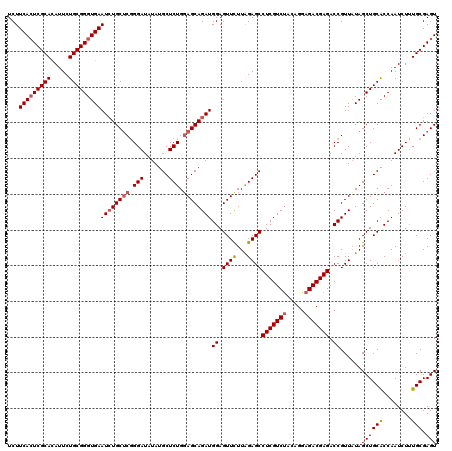
| Location | 1,297,363 – 1,297,483 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -40.15 |
| Consensus MFE | -35.04 |
| Energy contribution | -35.60 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1297363 120 - 22224390 CUCCUUCAAAACUUGCGGUGAUUUGUCUUGACUGGCAAUCUCUUCACUCGCACAUUCUGCGGGUGAAUCUGCUCUGGAUAUAUGCUCUGGAGCAGAUGGAGUUCUUAGAGCCUCGUCUAA ((((............((.((((.(((......))))))).))(((((((((.....)))))))))((((((((..((.......))..))))))))))))....((((......)))). ( -41.70) >DroSec_CAF1 34368 120 - 1 CUCCUUCGAAACUUGUGGUGAUUUGUCUUGACUGGCAAUCUCUUCACUCGCAUAUUCUGCGGGUGAAUGUGCUCGGGAUAUAUGCUCUGUAGCAGAUGGAGUUCUUAGAGCCUCGUCUAC ............(((..(.(...((((((((...(((.....((((((((((.....))))))))))..)))))))))))....).)..))).(((((..((((...))))..))))).. ( -35.40) >DroSim_CAF1 23231 120 - 1 CUCCUUCGAAACUUGUGGUGAUUUGUCUUGACUGGCAAUCUCUUCACUCGCAUAUUCUGCGGGUGAAUCUGCUCGGGAUAUAUGCUCUGGAGCAGAUGGAGUUCUUGGAGCCUCGUCUAC ...(((((((((((..((.((((.(((......))))))).))(((((((((.....)))))))))((((((((.(((.......))).)))))))).))))).)))))).......... ( -42.40) >DroYak_CAF1 30916 120 - 1 CUCCUUCGAAGCUUGUGGUGAUAUGUCAUCACUGGCAAUCUCUUCAGUCGCACAUUCUGCGGGUGAAUCUGCACGGGAUAUAUGCUCUGGAGCAGAUGGAGUUUUUGGAGCCUCGUCCAC ...((((((((((.(((((((....))))))).)))...(((((((.(((((.....))))).)))((((((.(.(((.......))).).))))))))))..))))))).......... ( -41.10) >consensus CUCCUUCGAAACUUGUGGUGAUUUGUCUUGACUGGCAAUCUCUUCACUCGCACAUUCUGCGGGUGAAUCUGCUCGGGAUAUAUGCUCUGGAGCAGAUGGAGUUCUUAGAGCCUCGUCUAC ((((............((.(((.((((......))))))).))(((((((((.....)))))))))((((((((.(((.......))).))))))))))))....((((......)))). (-35.04 = -35.60 + 0.56)

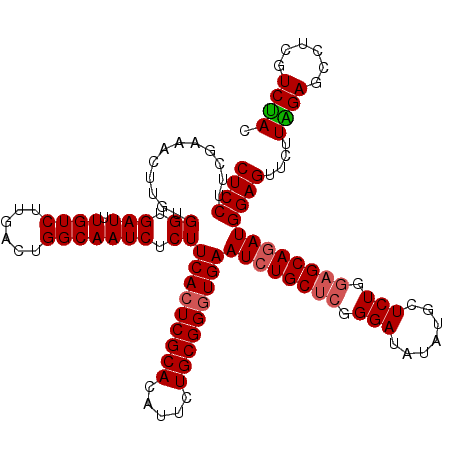
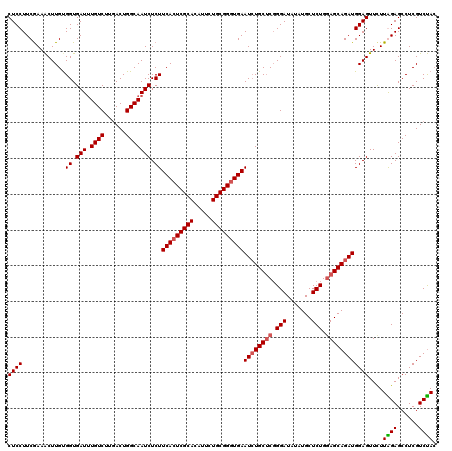
| Location | 1,297,403 – 1,297,523 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -32.57 |
| Energy contribution | -32.13 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1297403 120 - 22224390 CGUCAUCCUUGCUUUCUGCGACCCACGCUGGUCUUGAGGACUCCUUCAAAACUUGCGGUGAUUUGUCUUGACUGGCAAUCUCUUCACUCGCACAUUCUGCGGGUGAAUCUGCUCUGGAUA ....(((((((((.((.((((..(((((.(((.((((((....)))))).))).)).)))..))))...))..)))))....((((((((((.....))))))))))........)))). ( -38.90) >DroSec_CAF1 34408 120 - 1 CGUCAUCUUUGCUUUCUGUAACCGACGCUGGUCGUGAGGACUCCUUCGAAACUUGUGGUGAUUUGUCUUGACUGGCAAUCUCUUCACUCGCAUAUUCUGCGGGUGAAUGUGCUCGGGAUA .(((....((((.....))))(((((((..((((.((.((.((((.(((...))).)).)).)).)).))))..))......((((((((((.....))))))))))...).))))))). ( -38.10) >DroSim_CAF1 23271 120 - 1 CGUCAUCUUUGCUUUCUGUAACAGACGCUGGUCGUGAGGACUCCUUCGAAACUUGUGGUGAUUUGUCUUGACUGGCAAUCUCUUCACUCGCAUAUUCUGCGGGUGAAUCUGCUCGGGAUA .............(((((...((((.((..((((.((.((.((((.(((...))).)).)).)).)).))))..))......((((((((((.....))))))))))))))..))))).. ( -37.60) >DroYak_CAF1 30956 120 - 1 CACCAUCUUUGCUUUCUGCAACCGACGCUGGUCUUGAGGACUCCUUCGAAGCUUGUGGUGAUAUGUCAUCACUGGCAAUCUCUUCAGUCGCACAUUCUGCGGGUGAAUCUGCACGGGAUA ....(((.((((.....))))(((..((.(((.((((((....)))))).(((.(((((((....))))))).))).......(((.(((((.....))))).)))))).)).)))))). ( -36.20) >consensus CGUCAUCUUUGCUUUCUGCAACCGACGCUGGUCGUGAGGACUCCUUCGAAACUUGUGGUGAUUUGUCUUGACUGGCAAUCUCUUCACUCGCACAUUCUGCGGGUGAAUCUGCUCGGGAUA .((((((.((((.....)))).....((.(((..(((((....)))))..))).)))))))).((((......))))(((((((((((((((.....)))))))))).......))))). (-32.57 = -32.13 + -0.44)

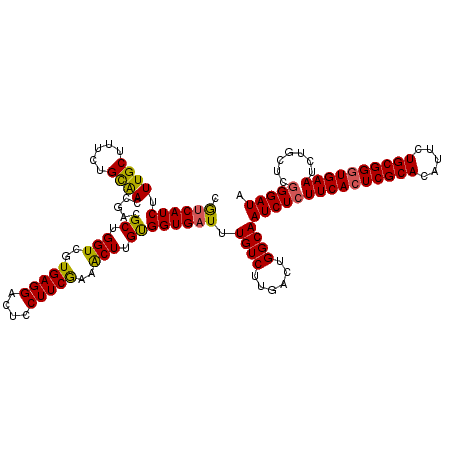
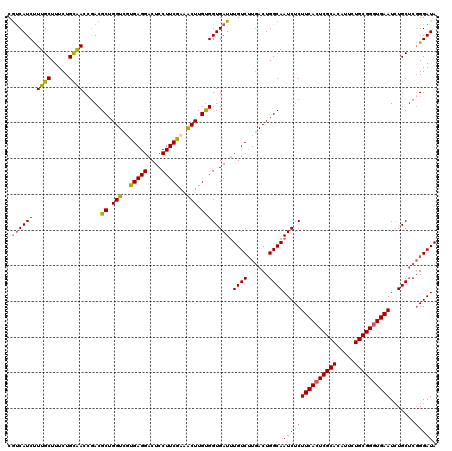
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:41 2006