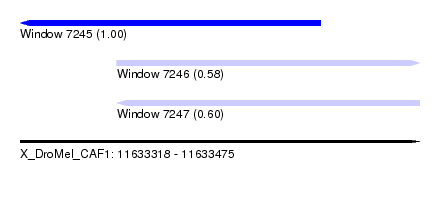
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,633,318 – 11,633,475 |
| Length | 157 |
| Max. P | 0.995878 |

| Location | 11,633,318 – 11,633,436 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -33.44 |
| Energy contribution | -32.92 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11633318 118 - 22224390 GGAUAUUGAUCUGAACCGUGAAUGUCUAUACCAAAUGAGUUGGCCGAGCCAACGGAAAAGUUCCAUGGAAAAUCCGGGAAAACGGAGCGGUGAUAUGAGUGUCCUUCUAAAUCUCCAA (((((((.((....(((((...........(((.....((((((...))))))(((.....))).)))....((((......)))))))))...)).))))))).............. ( -35.30) >DroSec_CAF1 6594 118 - 1 GGAUAUUGAUCUGAACCGUGAAUGUCUAUACCAAAUGAGUUGGCCGAGCCAACGGAAAAGUUCCAUGGGAAAUCCGGGAAAACGGAGCGGUGAUAUGAGUGUCCUUCUAAAUCUCCAA (((((((.((....(((((...........(((.....((((((...))))))(((.....))).)))....((((......)))))))))...)).))))))).............. ( -35.50) >DroSim_CAF1 5903 118 - 1 GGAUAUUGAUCUGAACCGUGAAUGUCUAUACCAAAUGAGUUGGCCGAGCCAACGGAAAAGUUCCAUGGGAAAUCCGGGAAAACGGAGCGGUGAUAUGAGUGUCCUUCUAAAUCUCCAA (((((((.((....(((((...........(((.....((((((...))))))(((.....))).)))....((((......)))))))))...)).))))))).............. ( -35.50) >DroEre_CAF1 7145 118 - 1 GGAUAUUAGUCUGAACCGUGAAUAACUAUACCAAAUGAGUUGGCCGAGUCAAUGAAAAAGUUCCAUGGGAUAUCCGGGGGAACGGAGCGGUGUUAUGAAUGUCCUUCAAAAUCUCCAA (((((((.......(((((.....(((...((((.....))))...)))..........(((((.(.((....)).).)))))...)))))......))))))).............. ( -29.12) >DroYak_CAF1 5933 118 - 1 GGAUAUUGAUCUGAACCGUGAAUGUCUAUACCAAAUGAGUUGGCCGAGCCAACGGAAAAGUUCCAUGGGAAAUCCGGGGGAACGGAGCGGUGAUAUGAAUGUCCUUCUAAAUCUCCAA (((((((.((....(((((...(((((...((......((((((...))))))(((....((((...)))).))))).)).)))..)))))...)).))))))).............. ( -35.60) >consensus GGAUAUUGAUCUGAACCGUGAAUGUCUAUACCAAAUGAGUUGGCCGAGCCAACGGAAAAGUUCCAUGGGAAAUCCGGGAAAACGGAGCGGUGAUAUGAGUGUCCUUCUAAAUCUCCAA (((((((.((....(((((...........(((.....((((((...))))))(((.....))).)))....((((......)))))))))...)).))))))).............. (-33.44 = -32.92 + -0.52)

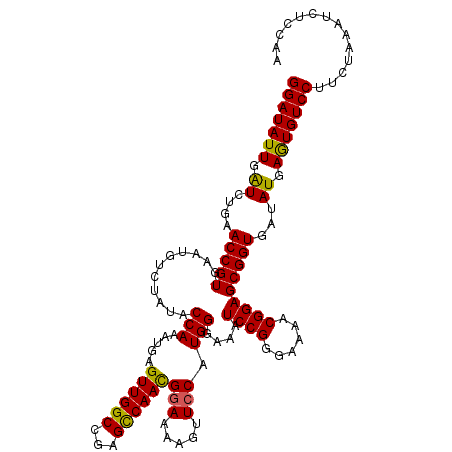

| Location | 11,633,356 – 11,633,475 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.79 |
| Mean single sequence MFE | -22.86 |
| Consensus MFE | -19.42 |
| Energy contribution | -19.46 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11633356 119 + 22224390 UUCCCGGAUUUUCCAUGGAACUUUUCCGUUGGCUCGGCCAACUCAUUUGGUAUAGACAUUCACGGUUCAGAUCAAUAUCCUGAUGCCAU-UCCAUGCCAUUAUAAUCUCAACCCGUUUUC ....(((.........(((.....)))((((((...)))))).....((((((.(.....)..(((((((.........)))).)))..-...)))))).............)))..... ( -23.70) >DroSec_CAF1 6632 119 + 1 UUCCCGGAUUUCCCAUGGAACUUUUCCGUUGGCUCGGCCAACUCAUUUGGUAUAGACAUUCACGGUUCAGAUCAAUAUCCUGAUGCCAU-UCCAUGCCAUUAUAAUCUCAACCCGUUUUC ....(((.........(((.....)))((((((...)))))).....((((((.(.....)..(((((((.........)))).)))..-...)))))).............)))..... ( -23.70) >DroSim_CAF1 5941 119 + 1 UUCCCGGAUUUCCCAUGGAACUUUUCCGUUGGCUCGGCCAACUCAUUUGGUAUAGACAUUCACGGUUCAGAUCAAUAUCCUGAUGCCAU-UCCAUGCCAUUAUAAUCUCAACCCGUUUUC ....(((.........(((.....)))((((((...)))))).....((((((.(.....)..(((((((.........)))).)))..-...)))))).............)))..... ( -23.70) >DroEre_CAF1 7183 116 + 1 CCCCCGGAUAUCCCAUGGAACUUUUUCAUUGACUCGGCCAACUCAUUUGGUAUAGUUAUUCACGGUUCAGACUAAUAUCCAGAAGCCAU-UCCAUACCAUUUUCAACUCAAACCGCC--- .....((((((......(((((.......(((((..(((((.....)))))..))))).....)))))......)))))).........-...........................--- ( -19.70) >DroYak_CAF1 5971 120 + 1 CCCCCGGAUUUCCCAUGGAACUUUUCCGUUGGCUCGGCCAACUCAUUUGGUAUAGACAUUCACGGUUCAGAUCAAUAUCCUGAAGCCAUUUCCAUACCAUUAUAAUCUCAAAUCGUUUUC .....(((.((((...))))....)))((((((...)))))).....((((((.((.((....(((((((.........))).)))))).)).))))))..................... ( -23.50) >consensus UUCCCGGAUUUCCCAUGGAACUUUUCCGUUGGCUCGGCCAACUCAUUUGGUAUAGACAUUCACGGUUCAGAUCAAUAUCCUGAUGCCAU_UCCAUGCCAUUAUAAUCUCAACCCGUUUUC ....(((.........(((.....)))((((((...)))))).....((((((.(.....)..(((((((.........)))).)))......)))))).............)))..... (-19.42 = -19.46 + 0.04)

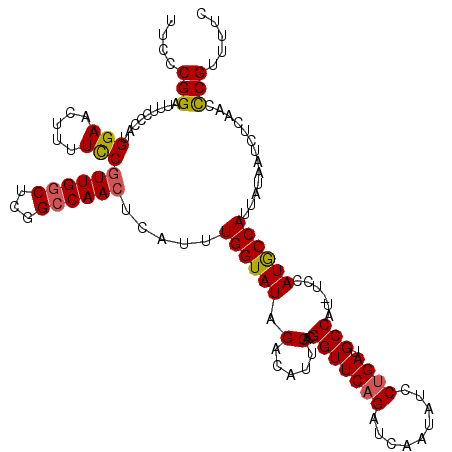
| Location | 11,633,356 – 11,633,475 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.79 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -24.10 |
| Energy contribution | -24.22 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11633356 119 - 22224390 GAAAACGGGUUGAGAUUAUAAUGGCAUGGA-AUGGCAUCAGGAUAUUGAUCUGAACCGUGAAUGUCUAUACCAAAUGAGUUGGCCGAGCCAACGGAAAAGUUCCAUGGAAAAUCCGGGAA .....((((((...........(((((...-((((..(((((.......))))).))))..)))))....(((.....((((((...))))))(((.....))).)))..)))))).... ( -32.80) >DroSec_CAF1 6632 119 - 1 GAAAACGGGUUGAGAUUAUAAUGGCAUGGA-AUGGCAUCAGGAUAUUGAUCUGAACCGUGAAUGUCUAUACCAAAUGAGUUGGCCGAGCCAACGGAAAAGUUCCAUGGGAAAUCCGGGAA .....((((((...........(((((...-((((..(((((.......))))).))))..)))))....(((.....((((((...))))))(((.....))).)))..)))))).... ( -33.00) >DroSim_CAF1 5941 119 - 1 GAAAACGGGUUGAGAUUAUAAUGGCAUGGA-AUGGCAUCAGGAUAUUGAUCUGAACCGUGAAUGUCUAUACCAAAUGAGUUGGCCGAGCCAACGGAAAAGUUCCAUGGGAAAUCCGGGAA .....((((((...........(((((...-((((..(((((.......))))).))))..)))))....(((.....((((((...))))))(((.....))).)))..)))))).... ( -33.00) >DroEre_CAF1 7183 116 - 1 ---GGCGGUUUGAGUUGAAAAUGGUAUGGA-AUGGCUUCUGGAUAUUAGUCUGAACCGUGAAUAACUAUACCAAAUGAGUUGGCCGAGUCAAUGAAAAAGUUCCAUGGGAUAUCCGGGGG ---...(..(((.((..(...((((((((.-((((.(((.((((....)))))))))))......))))))))......)..)))))..)............((.(((.....))).)). ( -26.40) >DroYak_CAF1 5971 120 - 1 GAAAACGAUUUGAGAUUAUAAUGGUAUGGAAAUGGCUUCAGGAUAUUGAUCUGAACCGUGAAUGUCUAUACCAAAUGAGUUGGCCGAGCCAACGGAAAAGUUCCAUGGGAAAUCCGGGGG .....................(((((((((.((((.((((((.......)))))))))).....))))))))).....((((((...))))))(((....((((...)))).)))..... ( -33.40) >consensus GAAAACGGGUUGAGAUUAUAAUGGCAUGGA_AUGGCAUCAGGAUAUUGAUCUGAACCGUGAAUGUCUAUACCAAAUGAGUUGGCCGAGCCAACGGAAAAGUUCCAUGGGAAAUCCGGGAA .....((((((.........(((((((....((((.....((((....))))...))))..))).)))).(((.....((((((...))))))(((.....))).)))..)))))).... (-24.10 = -24.22 + 0.12)

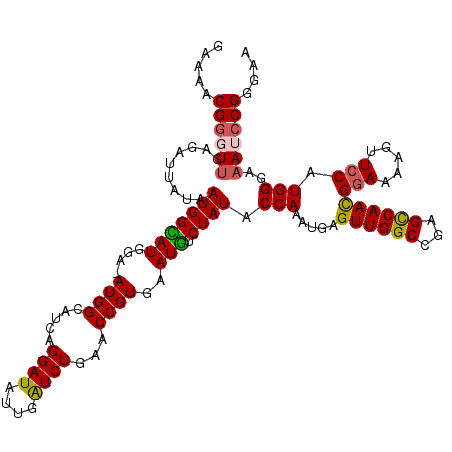
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:22 2006