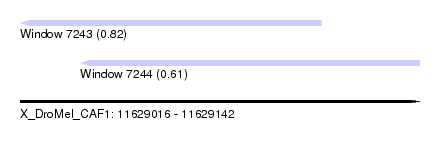
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,629,016 – 11,629,142 |
| Length | 126 |
| Max. P | 0.818519 |

| Location | 11,629,016 – 11,629,111 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -23.04 |
| Consensus MFE | -19.67 |
| Energy contribution | -19.28 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
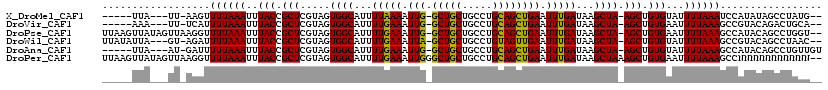
>X_DroMel_CAF1 11629016 95 - 22224390 GCAUUUUAAAAUUGGCUGCUGCCUGCAGCUGAAUUUGAUAAGCUAAGCUGUGUAUUUUAAAUCCAUAUAGCCUAUGU-AUGUGAUGAUCA------------AGUUGA ((...(((((.(..(((((.....)))))..).)))))...))..((((.((.(((....(((((((((.....)))-))).))))))))------------)))).. ( -22.60) >DroPse_CAF1 2930 91 - 1 GCAUUUUGAAAUUGGCUGCUGCCUGCAGCUGAAUUUGAUAAGCUAAGCUGUGAAUUUUAAAGCCAUACAGCCUGGUU-UU-U---GGUUC------------AUUGGU ((...(..((.(..(((((.....)))))..).))..)...))....(.(((((((..(((((((.......)))))-))-.---)))))------------)).).. ( -26.70) >DroGri_CAF1 5144 107 - 1 GCAUUUUGAAAUUGGCUGCUGCCUGCAGCUGAAUUUGAUAAGCUAAGCUGUGAAUUUUAAAGCCGUACAGACUGAAUUAU-CGUUGUUUAUAUAGGUGCAACAGUUGU ((((((.(((.(..(((((.....)))))..).)))(((((....(((((((.............))))).))...))))-)...........))))))......... ( -24.82) >DroEre_CAF1 1935 94 - 1 GCAUUUUAAAAUUGGCUGCUGCCUGCAGCUGAAUUUGAUAAGCUAAGCUGUGUAUUUUAAAGCCAUACAGCCUAUUU-GU-UGUUGUUUA------------AGUUGU (((.((((((.(..(((((.....)))))..)....((((((.((.(((((((...........))))))).)))))-))-)....))))------------)).))) ( -23.70) >DroWil_CAF1 9310 91 - 1 GCAUUUUGAAAUUAGCUGCUGCCUGUAGUUGAAUUUGAUAAGCUAAGCUGUGUAUUUUAAAGCCGUACAGCCUAACG-AA-G---GACGA------------AGAAGA ((...(..((.((((((((.....)))))))).))..)...))...((((((.............))))))....(.-..-.---)....------------...... ( -17.62) >DroYak_CAF1 1642 86 - 1 GCAUUUUAAAAUUGGCUGCUGCCUGCAGCUGAAUUUGAUAAGCUAAGCUGUGUAUUUUAAAGCCAUACAGCCUAUUU-GU-UGUUGUU-------------------- ((...(((((.(..(((((.....)))))..).)))))...))...(((((((...........)))))))......-..-.......-------------------- ( -22.80) >consensus GCAUUUUAAAAUUGGCUGCUGCCUGCAGCUGAAUUUGAUAAGCUAAGCUGUGUAUUUUAAAGCCAUACAGCCUAUUU_AU_UGUUGUUUA____________AGUUGU ((...(((((.((((((((.....)))))))).)))))...))...(((((((...........)))))))..................................... (-19.67 = -19.28 + -0.39)


| Location | 11,629,035 – 11,629,142 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.56 |
| Mean single sequence MFE | -28.89 |
| Consensus MFE | -19.85 |
| Energy contribution | -19.30 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11629035 107 - 22224390 -----UUA---UU-AAGUUUUAAAUUUACCGCUCGUAGUGGCAUUUUAAAAUUG-GCUGCUGCCUGCAGCUGAAUUUGAUAAGCUA-AGCUGUGUAUUUUAAAUCCAUAUAGCCUAUG-- -----(((---((-((((((((((....((((.....))))...)))))).(..-(((((.....)))))..))))))))))....-.(((((((...........))))))).....-- ( -26.60) >DroVir_CAF1 5404 107 - 1 -----AAA---UU-UCAUUUUAAAUUUACCGCUCGUAGUGGCAUUUUGAAAUUG-GCUGCUGCCUGCAGCUGAAUUUGAUAAGCUA-AGCUGUGAAUUUUAAAGCCGUACAGACUGCA-- -----...---..-...(((.((((((((.(((.....((((...(..((.(..-(((((.....)))))..).))..)...))))-))).)))))))).)))((.((....)).)).-- ( -28.60) >DroPse_CAF1 2945 116 - 1 UUAAGUUAUAGUUAAGGUUUUAAAUUUACCGCUCGUAGUGGCAUUUUGAAAUUG-GCUGCUGCCUGCAGCUGAAUUUGAUAAGCUA-AGCUGUGAAUUUUAAAGCCAUACAGCCUGGU-- ...............((((((((((((((.(((.....((((...(..((.(..-(((((.....)))))..).))..)...))))-))).))))).)))))))))............-- ( -33.50) >DroWil_CAF1 9325 112 - 1 UUAUAUUA---GU-AGAUUUUAAAUUUACCGCUCGUAGUGGCAUUUUGAAAUUA-GCUGCUGCCUGUAGUUGAAUUUGAUAAGCUA-AGCUGUGUAUUUUAAAGCCGUACAGCCUAAC-- .....(((---((-.....((((((((((..(..((((..((............-))..))))..)..).)))))))))...))))-)((((((.............)))))).....-- ( -25.12) >DroAna_CAF1 1631 109 - 1 -----UUA---AU-GAUUUUUAAAUUUACCGCUCGUAGUGGCAUUUUGAAAUUG-GCUGCUGCCUGCAGCUGAAUUUGAUAAGCUA-AGCUGUGUAUUUUAAAGCCAUACAGCCUGUUGU -----...---..-...(((((((....((((.....))))...)))))))(..-(((((.....)))))..)........(((..-.(((((((...........)))))))..))).. ( -26.20) >DroPer_CAF1 2018 118 - 1 UUAAGUUAUAGUUAAGGUUUUAAAUUUACCGCUCGUAGUGGCAUUUUGAAAUUGGGCUGCUGCCUGCAGCUGAAUUUGAUAAGCUAAAGCUGUGAAUUUUAAAGCCNNNNNNNNNNNN-- ...............((((((((((((((.(((.....((((.((.(.(((((.((((((.....)))))).))))).).)))))).))).))))).)))))))))............-- ( -33.30) >consensus _____UUA___UU_AGGUUUUAAAUUUACCGCUCGUAGUGGCAUUUUGAAAUUG_GCUGCUGCCUGCAGCUGAAUUUGAUAAGCUA_AGCUGUGAAUUUUAAAGCCAUACAGCCUGUU__ ..................((((((..(((.(((.....((((...(((((.(((.(((((.....)))))))).)))))...)))).))).)))...))))))................. (-19.85 = -19.30 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:19 2006