
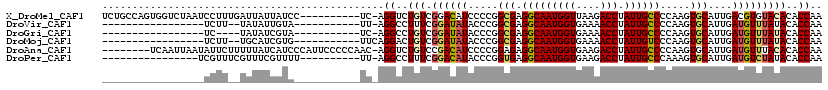
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,628,717 – 11,628,826 |
| Length | 109 |
| Max. P | 0.981966 |

| Location | 11,628,717 – 11,628,826 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.03 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -15.87 |
| Energy contribution | -16.90 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11628717 109 + 22224390 UCUGCCAGUGGUCUAAUCCUUUGAUUAUUAUCC----------UC-AGGUCUGUCGGACAUCCCCGGCGAGGCAAUGGUUAAGACCUAUUGCCCCAAGUGCAUUGACGUGUACACACCAA ...(((.(.((..(((((....)))))....))----------.)-.)))..(((((......)))))(.(((((((((....))).)))))).)..((((((....))))))....... ( -30.30) >DroVir_CAF1 4328 89 + 1 -----------------UCUU--UAUAUUGUA-----------UU-AGGCCUUUCGGAUAUACCCGGCGAGGCAAUGGUGAAAACCUAUUGCCCCAAGUGCAUUGAUGUUUAUACACCAA -----------------....--.....((((-----------((-.((....((((......))))...(((((((((....))).)))))))).))))))(((.(((....))).))) ( -23.80) >DroGri_CAF1 1297 87 + 1 -----------------UC----UAUAUCGUA-----------UC-AGGCCUGUCGGAUAUACCCGGCGAGGCAAUGGUGAAAACCUAUUGCCCCAAGUGCAUUGAUGUUUAUACACCAA -----------------..----((((...((-----------((-(((((((((((......)))))(.(((((((((....))).)))))).).)).)).))))))..))))...... ( -28.30) >DroMoj_CAF1 1101 90 + 1 -----------------UCUU--UGCAUCGUG-----------UUCAGGACUGUCGGAUAUACCCGGCGAGGCAAUGGUGAAAACCUAUUGUCCCAAGUGCAUUGAUGUUUAUACACCAA -----------------....--.((((((((-----------(...((..((((((......)))))).(((((((((....))).))))))))....))).))))))........... ( -26.70) >DroAna_CAF1 1252 111 + 1 --------UCAAUUAAUAUUCUUUUUAUCAUCCCAUUCCCCCAAC-AGGUCUGUCCGACAUCCCCGGAGAGGCAAUGGUGAAGACCUAUUGCCCCAAGUGCAUUGAUGUUUACACACCAA --------.....................................-.(((.(((..((((((........(((((((((....))).))))))...........)))))).))).))).. ( -20.41) >DroPer_CAF1 1590 93 + 1 ----------------UCGUUUCGUUUCGUUUU----------UU-AGGCCUUUCGGACAUACCCGGUGAGGCAAUGGUGAAGACCUAUUGCCCAAAGUGCAUUGAUGUCUAUACACCAA ----------------.................----------..-..(((((((((......)))).)))))..(((((.((((....(((.......))).....))))...))))). ( -22.90) >consensus _________________UCUU__UUUAUCGUA___________UC_AGGCCUGUCGGACAUACCCGGCGAGGCAAUGGUGAAAACCUAUUGCCCCAAGUGCAUUGAUGUUUAUACACCAA ...............................................((..(((.((((((.....(((.(((((((((....))).)))))).....)))....)))))))))..)).. (-15.87 = -16.90 + 1.03)
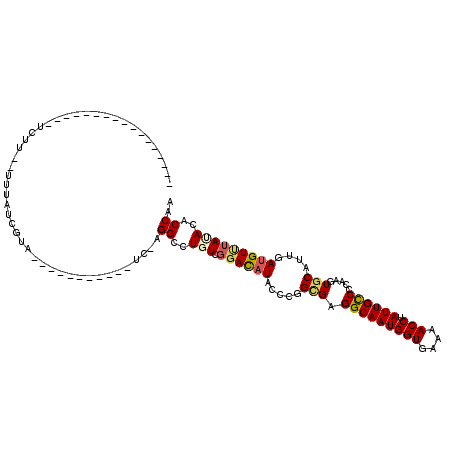

| Location | 11,628,717 – 11,628,826 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.03 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -20.06 |
| Energy contribution | -20.33 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11628717 109 - 22224390 UUGGUGUGUACACGUCAAUGCACUUGGGGCAAUAGGUCUUAACCAUUGCCUCGCCGGGGAUGUCCGACAGACCU-GA----------GGAUAAUAAUCAAAGGAUUAGACCACUGGCAGA ..(((((((........))))))).((((((((.(((....)))))))))))(((((((.(((((..((....)-).----------))))).(((((....)))))..)).)))))... ( -38.50) >DroVir_CAF1 4328 89 - 1 UUGGUGUAUAAACAUCAAUGCACUUGGGGCAAUAGGUUUUCACCAUUGCCUCGCCGGGUAUAUCCGAAAGGCCU-AA-----------UACAAUAUA--AAGA----------------- ..(((((((........))))))).((((((((.(((....))))))))))).....((((..((....))...-.)-----------)))......--....----------------- ( -25.60) >DroGri_CAF1 1297 87 - 1 UUGGUGUAUAAACAUCAAUGCACUUGGGGCAAUAGGUUUUCACCAUUGCCUCGCCGGGUAUAUCCGACAGGCCU-GA-----------UACGAUAUA----GA----------------- ..(((((((........))))))).((((((((.(((....)))))))))))(((.((.....))....)))..-..-----------.........----..----------------- ( -24.90) >DroMoj_CAF1 1101 90 - 1 UUGGUGUAUAAACAUCAAUGCACUUGGGACAAUAGGUUUUCACCAUUGCCUCGCCGGGUAUAUCCGACAGUCCUGAA-----------CACGAUGCA--AAGA----------------- (((((((....)))))))((((((..((((....(((....)))..(((((....))))).........))))..).-----------...).))))--....----------------- ( -22.00) >DroAna_CAF1 1252 111 - 1 UUGGUGUGUAAACAUCAAUGCACUUGGGGCAAUAGGUCUUCACCAUUGCCUCUCCGGGGAUGUCGGACAGACCU-GUUGGGGGAAUGGGAUGAUAAAAAGAAUAUUAAUUGA-------- (((((((....)))))))(((.(....))))....(((.((.(((((.((((..((((..((.....))..)))-)..)))).))))))).)))..................-------- ( -34.60) >DroPer_CAF1 1590 93 - 1 UUGGUGUAUAGACAUCAAUGCACUUUGGGCAAUAGGUCUUCACCAUUGCCUCACCGGGUAUGUCCGAAAGGCCU-AA----------AAAACGAAACGAAACGA---------------- (((((((....))))))).((.(((((((((((.(((....))))))))))...((((....))))))))))..-..----------.................---------------- ( -23.30) >consensus UUGGUGUAUAAACAUCAAUGCACUUGGGGCAAUAGGUCUUCACCAUUGCCUCGCCGGGUAUAUCCGACAGGCCU_AA___________GACGAUAAA__AAGA_________________ ..(((((((........))))))).((((((((.(((....)))))))))))..((((....))))...................................................... (-20.06 = -20.33 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:17 2006