| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,626,343 – 11,626,543 |
| Length | 200 |
| Max. P | 0.875727 |

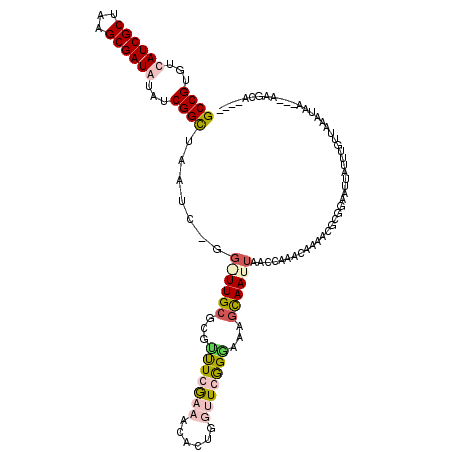
| Location | 11,626,343 – 11,626,463 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.26 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -15.20 |
| Energy contribution | -17.45 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.51 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11626343 120 - 22224390 GCCGUGUCAUCGCUAAGCGAUGUAUCGGCUAAUCGGGAUUGCGCGUUUCGCAACACUGGUUCGGAAAAGCAAUUAACCAAACAAAACGCGGAAUUAUUUGUUAAAUAAGCAAAUCAAGCC ((((...((((((...))))))...)))).....((((((((...(((((.(((....))))))))..))))))..))...........((....(((((((.....)))))))....)) ( -33.70) >DroSim_CAF1 56249 117 - 1 GCCGUGUCAUCGCUAAGCGAUGUAUCGGCUAAUCAGGGUUGCGCGUUUCGCAACACUGGUUCGGGAAAGUAAUUAACCAAACAAAACGCGGAAUUAUUUCUUAAAUAA---AAGCAAGCC ((((...((((((...))))))...))))......((.((((((((((..((....))(((.((..((....))..)).))).))))))((((....)))).......---..)))).)) ( -34.80) >DroEre_CAF1 57106 97 - 1 GCCGUGUCAUCGCUAAGCGAUAUAUCGGCAAAU-------------CUGGAAACACUGUUUCGGGAACG---UUAUACUAAUAUAAGUAGGAAUGAUUUGUCAAAUAA---AUGAA---- (((((((.(((((...)))))))).))))...(-------------((.(((((...))))).))).((---((.((((......))))..)))).............---.....---- ( -23.60) >DroYak_CAF1 58524 113 - 1 GCCGUGUCAUCGCUAAGCGAUAUAUCGGUUUGUAUGGCUUGCGCGCUUGAAAACACUGUUUCAGGAAAGCAAUUAACCUAAUAUAACGUGGCAUUAUUUGUUAAAAAA---AUGCA---- (((((((....((.((((.(((((......))))).))))))((.(((((((......)))))))...))...............)))))))................---.....---- ( -27.80) >consensus GCCGUGUCAUCGCUAAGCGAUAUAUCGGCUAAUC_GG_UUGCGCGUUUCGAAACACUGGUUCGGGAAAGCAAUUAACCAAACAAAACGCGGAAUUAUUUGUUAAAUAA___AAGCA____ ((((...((((((...))))))...)))).......((((((...(((((((......)))))))...)))))).............................................. (-15.20 = -17.45 + 2.25)


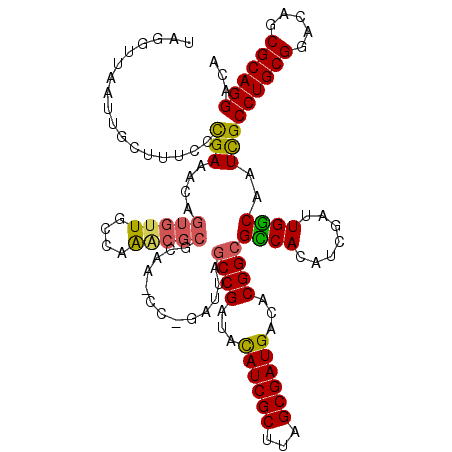
| Location | 11,626,383 – 11,626,503 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -38.41 |
| Consensus MFE | -28.77 |
| Energy contribution | -28.65 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11626383 120 + 22224390 UUGGUUAAUUGCUUUUCCGAACCAGUGUUGCGAAACGCGCAAUCCCGAUUAGCCGAUACAUCGCUUAGCGAUGACACGGCGCCACAUCGAUUGGCAAUCGCCUGCGGACAGCGCAGGACA .(((((.............)))))(..(((((.....)))))..)(((((.((((...((((((...))))))...))))((((.......)))))))))((((((.....))))))... ( -42.42) >DroSim_CAF1 56286 120 + 1 UUGGUUAAUUACUUUCCCGAACCAGUGUUGCGAAACGCGCAACCCUGAUUAGCCGAUACAUCGCUUAGCGAUGACACGGCGCCACAUCGAUUGGCAAUUGCCUGCGGACAGCGCAGGACA ((((..((....))..))))..(((.((((((.....)))))).)))....((((...((((((...))))))...))))((((.......))))...((((((((.....)))))).)) ( -44.00) >DroEre_CAF1 57139 104 + 1 UAGUAUAA---CGUUCCCGAAACAGUGUUUCCAG-------------AUUUGCCGAUAUAUCGCUUAGCGAUGACACGGCGUCACAUCGAUUGACAAUCGCCUGCGGACAGCGCAGGACG ........---.......(((((...)))))..(-------------(((.((((...((((((...))))))...))))((((.......))))))))(((((((.....)))))).). ( -29.80) >DroYak_CAF1 58557 120 + 1 UAGGUUAAUUGCUUUCCUGAAACAGUGUUUUCAAGCGCGCAAGCCAUACAAACCGAUAUAUCGCUUAGCGAUGACACGGCGUCACGGCGAUUGGCAAUCGCCUGCGGACAGCGCAGGACA ...((((((((((....(((....(((((.((((((((....))..........((....))))))...)).)))))....))).))))))))))....(((((((.....)))))).). ( -37.40) >consensus UAGGUUAAUUGCUUUCCCGAAACAGUGUUGCCAAACGCGCAA_CC_GAUUAGCCGAUACAUCGCUUAGCGAUGACACGGCGCCACAUCGAUUGGCAAUCGCCUGCGGACAGCGCAGGACA .................(((....(((((....))))).............((((...((((((...))))))...))))((((.......))))..)))((((((.....))))))... (-28.77 = -28.65 + -0.12)


| Location | 11,626,383 – 11,626,503 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -42.38 |
| Consensus MFE | -28.90 |
| Energy contribution | -30.77 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11626383 120 - 22224390 UGUCCUGCGCUGUCCGCAGGCGAUUGCCAAUCGAUGUGGCGCCGUGUCAUCGCUAAGCGAUGUAUCGGCUAAUCGGGAUUGCGCGUUUCGCAACACUGGUUCGGAAAAGCAAUUAACCAA .(.((((((.....)))))))((((((((.......))))((((...((((((...))))))...)))).))))((((((((...(((((.(((....))))))))..))))))..)).. ( -46.10) >DroSim_CAF1 56286 120 - 1 UGUCCUGCGCUGUCCGCAGGCAAUUGCCAAUCGAUGUGGCGCCGUGUCAUCGCUAAGCGAUGUAUCGGCUAAUCAGGGUUGCGCGUUUCGCAACACUGGUUCGGGAAAGUAAUUAACCAA ((.((((((.....))))))))...((((.......))))((((...((((((...))))))...)))).((((((.((((((.....)))))).)))))).((..((....))..)).. ( -47.40) >DroEre_CAF1 57139 104 - 1 CGUCCUGCGCUGUCCGCAGGCGAUUGUCAAUCGAUGUGACGCCGUGUCAUCGCUAAGCGAUAUAUCGGCAAAU-------------CUGGAAACACUGUUUCGGGAACG---UUAUACUA ((.((((((.....))))))))............(((((((((((((.(((((...)))))))).))))...(-------------((.(((((...))))).)))..)---)))))... ( -35.90) >DroYak_CAF1 58557 120 - 1 UGUCCUGCGCUGUCCGCAGGCGAUUGCCAAUCGCCGUGACGCCGUGUCAUCGCUAAGCGAUAUAUCGGUUUGUAUGGCUUGCGCGCUUGAAAACACUGUUUCAGGAAAGCAAUUAACCUA .(.((((((.....)))))))(((.....)))((((((..(((((((.(((((...)))))))).))))...))))))((((...(((((((......)))))))...))))........ ( -40.10) >consensus UGUCCUGCGCUGUCCGCAGGCGAUUGCCAAUCGAUGUGACGCCGUGUCAUCGCUAAGCGAUAUAUCGGCUAAUC_GG_UUGCGCGUUUCGAAACACUGGUUCGGGAAAGCAAUUAACCAA ((.((((((.....))))))))...((((.......))))((((...((((((...))))))...)))).......((((((...(((((((......)))))))...))))))...... (-28.90 = -30.77 + 1.88)


| Location | 11,626,423 – 11,626,543 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -40.47 |
| Consensus MFE | -32.92 |
| Energy contribution | -32.80 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.735316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11626423 120 + 22224390 AAUCCCGAUUAGCCGAUACAUCGCUUAGCGAUGACACGGCGCCACAUCGAUUGGCAAUCGCCUGCGGACAGCGCAGGACAGCGGUAAUCGAUAGUCGAGAUUGCAUACCUGAUAUCAUCG ((((.((((((((((...((((((...))))))...))))......(((((((.(.....((((((.....)))))).....).))))))))))))).)))).................. ( -42.40) >DroSim_CAF1 56326 120 + 1 AACCCUGAUUAGCCGAUACAUCGCUUAGCGAUGACACGGCGCCACAUCGAUUGGCAAUUGCCUGCGGACAGCGCAGGACAGCGGUAAUCGAUAGUCGAGAUUGCAUACCUGAUAUCAUCG ......(((((((((...((((((...))))))...))))......(((((((.(..(((((((((.....)))))).)))..)))))))))))))(((((..((....))..))).)). ( -39.30) >DroEre_CAF1 57170 113 + 1 -------AUUUGCCGAUAUAUCGCUUAGCGAUGACACGGCGUCACAUCGAUUGACAAUCGCCUGCGGACAGCGCAGGACGGCAGUUAUCGAUAGUGCCGAUUGCAUACCUGGUAUCAUCG -------....((((...((((((...))))))...))))(.(((((((((.(((..(((((((((.....)))))).)))..))))))))).))).)(((.(.((((...)))))))). ( -39.10) >DroYak_CAF1 58597 120 + 1 AAGCCAUACAAACCGAUAUAUCGCUUAGCGAUGACACGGCGUCACGGCGAUUGGCAAUCGCCUGCGGACAGCGCAGGACAGCGGUUAUCGAUAGUCGCGAUUGCAUACCUGGUACCAUCG ..((((..(((.(((...((((((...))))))...)))(((....))).)))(((((((((((((.....)))))).).(((..((....))..))))))))).....))))....... ( -41.10) >consensus AA_CC_GAUUAGCCGAUACAUCGCUUAGCGAUGACACGGCGCCACAUCGAUUGGCAAUCGCCUGCGGACAGCGCAGGACAGCGGUAAUCGAUAGUCGAGAUUGCAUACCUGAUAUCAUCG ...........((((...((((((...))))))...))))((((.......)))).....((((((.....)))))).(((..((((((.........))))))....)))......... (-32.92 = -32.80 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:15 2006