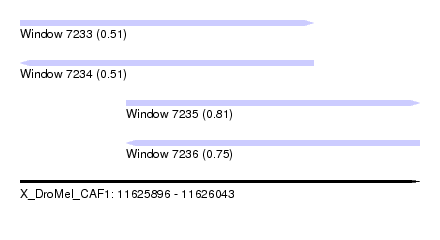
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,625,896 – 11,626,043 |
| Length | 147 |
| Max. P | 0.809108 |

| Location | 11,625,896 – 11,626,004 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.86 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -19.52 |
| Energy contribution | -21.50 |
| Covariance contribution | 1.98 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11625896 108 + 22224390 CAAAAUAACGUGUGUUUUGAAAAUAAACGGUAAU-AUAGCGCAGAAUAUAACAAAACAAUCGUACGCCCUGAGCGAGUUGAAGCGAUAAGGCC--UUG---------CUCUCGCUUUUUC .......(((..(((((((...(((....((...-.....))....)))..)))))))..))).......(((((((....(((((.......--)))---------))))))))).... ( -19.20) >DroSec_CAF1 56302 107 + 1 ----------UGUGCUUUGAAAAUAAACGGUAAUUAAAGCACAGGAUACAAAAAAACAA---UGCGCUCUGAGCGAGUUGAAGCGAUAAAGCAGGUUGCAAACUGCUCUCUCGCCUUGUC ----------((((((((((..((.....))..))))))))))....((((.....(((---(.(((.....))).))))..((((.(.(((((........))))).).)))).)))). ( -29.80) >DroSim_CAF1 55873 110 + 1 ----------UGUGCUUUGAAAAUAAACGGUAAUUAAAGCACAGGAUACAAAAAAACAAUCGUGCGCUCUGAGCGAGUUGAAGCGAUAAAGCAGGUUGCAAACUGCUCUCUCGCUUUGUC ----------((((((((((..((.....))..)))))))))).((((........((((....(((.....))).))))((((((.(.(((((........))))).).)))))))))) ( -29.60) >consensus __________UGUGCUUUGAAAAUAAACGGUAAUUAAAGCACAGGAUACAAAAAAACAAUCGUGCGCUCUGAGCGAGUUGAAGCGAUAAAGCAGGUUGCAAACUGCUCUCUCGCUUUGUC ..........((((((((((..((.....))..)))))))))).....................(((.....)))....(((((((.(.(((((........))))).).)))))))... (-19.52 = -21.50 + 1.98)

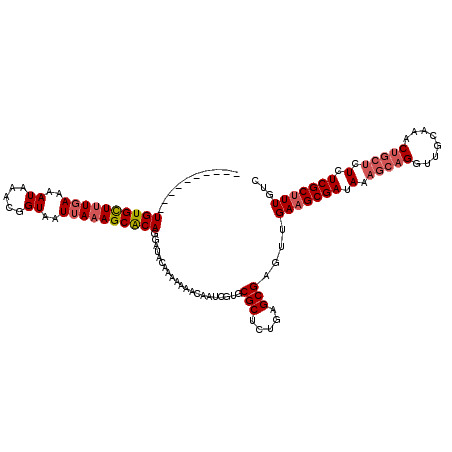
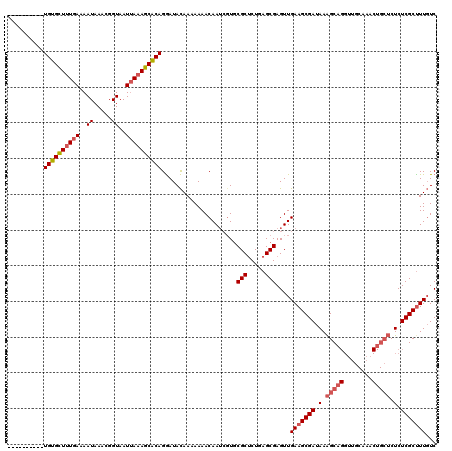
| Location | 11,625,896 – 11,626,004 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.86 |
| Mean single sequence MFE | -25.94 |
| Consensus MFE | -19.36 |
| Energy contribution | -20.57 |
| Covariance contribution | 1.21 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11625896 108 - 22224390 GAAAAAGCGAGAG---------CAA--GGCCUUAUCGCUUCAACUCGCUCAGGGCGUACGAUUGUUUUGUUAUAUUCUGCGCUAU-AUUACCGUUUAUUUUCAAAACACACGUUAUUUUG (((((((((((..---------..(--(((......))))...))))))...((((((.((.(((......))).))))))))..-...........))))).................. ( -22.60) >DroSec_CAF1 56302 107 - 1 GACAAGGCGAGAGAGCAGUUUGCAACCUGCUUUAUCGCUUCAACUCGCUCAGAGCGCA---UUGUUUUUUUGUAUCCUGUGCUUUAAUUACCGUUUAUUUUCAAAGCACA---------- .(((((((((.(((((((........))))))).))))).(((..(((.....)))..---))).....)))).....(((((((.................))))))).---------- ( -27.63) >DroSim_CAF1 55873 110 - 1 GACAAAGCGAGAGAGCAGUUUGCAACCUGCUUUAUCGCUUCAACUCGCUCAGAGCGCACGAUUGUUUUUUUGUAUCCUGUGCUUUAAUUACCGUUUAUUUUCAAAGCACA---------- ....((((((.(((((((........))))))).))))))......(((.((((((((.(((...........))).)))))))).......(........)..)))...---------- ( -27.60) >consensus GACAAAGCGAGAGAGCAGUUUGCAACCUGCUUUAUCGCUUCAACUCGCUCAGAGCGCACGAUUGUUUUUUUGUAUCCUGUGCUUUAAUUACCGUUUAUUUUCAAAGCACA__________ ....((((((.(((((((........))))))).))))))..........((((((((.(((...........))).)))))))).......((((.......))))............. (-19.36 = -20.57 + 1.21)

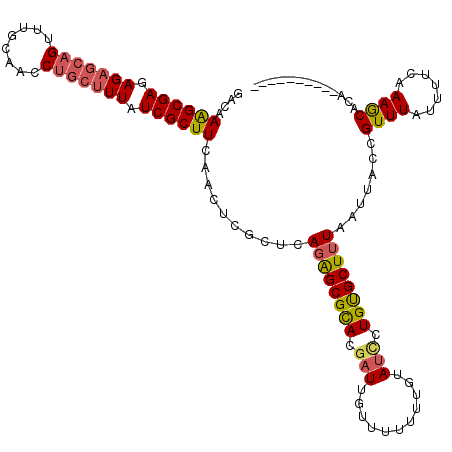

| Location | 11,625,935 – 11,626,043 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.75 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -25.45 |
| Energy contribution | -27.53 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11625935 108 + 22224390 GCAGAAUAUAACAAAACAAUCGUACGCCCUGAGCGAGUUGAAGCGAUAAGGCC--UUG---------CUCUCGCUUUUUCGCUUAAGCUCGCAACUCGCUUCGCUCUCACUCUGACAUC .((((..........((....))..((...((((((((((((((((.(((((.--...---------.....))))).)))))).......)))))))))).))......))))..... ( -28.81) >DroSec_CAF1 56332 116 + 1 ACAGGAUACAAAAAAACAA---UGCGCUCUGAGCGAGUUGAAGCGAUAAAGCAGGUUGCAAACUGCUCUCUCGCCUUGUCACCUGAGCUCGCAACUCGCUUCGCUCUCACUCUGACAAC ...................---.(((....((((((((((.(((.......(((((.((((...((......)).)))).))))).)))..)))))))))))))............... ( -31.30) >DroSim_CAF1 55903 119 + 1 ACAGGAUACAAAAAAACAAUCGUGCGCUCUGAGCGAGUUGAAGCGAUAAAGCAGGUUGCAAACUGCUCUCUCGCUUUGUCGCCUGAGCUCGCAACUCGCUUUGCUCUCACUCUGACAAC .((((((...........)))(((.((...((((((((((..(((((((((((((..((.....))..))).))))))))))..(....).)))))))))).))...))).)))..... ( -39.50) >consensus ACAGGAUACAAAAAAACAAUCGUGCGCUCUGAGCGAGUUGAAGCGAUAAAGCAGGUUGCAAACUGCUCUCUCGCUUUGUCGCCUGAGCUCGCAACUCGCUUCGCUCUCACUCUGACAAC .((((..........((....))..((...((((((((((..((((((((((.((..((.....))...)).)))))))))).........)))))))))).))......))))..... (-25.45 = -27.53 + 2.09)


| Location | 11,625,935 – 11,626,043 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.75 |
| Mean single sequence MFE | -38.17 |
| Consensus MFE | -33.09 |
| Energy contribution | -32.97 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11625935 108 - 22224390 GAUGUCAGAGUGAGAGCGAAGCGAGUUGCGAGCUUAAGCGAAAAAGCGAGAG---------CAA--GGCCUUAUCGCUUCAACUCGCUCAGGGCGUACGAUUGUUUUGUUAUAUUCUGC .....(((((((..((((((((((..((((..(((.(((((..(((((((((---------...--...))).))))))....))))).))).))))...)))))))))).))))))). ( -35.60) >DroSec_CAF1 56332 116 - 1 GUUGUCAGAGUGAGAGCGAAGCGAGUUGCGAGCUCAGGUGACAAGGCGAGAGAGCAGUUUGCAACCUGCUUUAUCGCUUCAACUCGCUCAGAGCGCA---UUGUUUUUUUGUAUCCUGU ((..((.((((((((((((((((.((((((((((..(....)...((......)))))))))))).))))...)))))....))))))).))..)).---................... ( -38.90) >DroSim_CAF1 55903 119 - 1 GUUGUCAGAGUGAGAGCAAAGCGAGUUGCGAGCUCAGGCGACAAAGCGAGAGAGCAGUUUGCAACCUGCUUUAUCGCUUCAACUCGCUCAGAGCGCACGAUUGUUUUUUUGUAUCCUGU .....(((..((.(((.(((((.(((((.(.((((.(((((..((((((.(((((((........))))))).))))))....)))))..)))).).))))))))))))).))..))). ( -40.00) >consensus GUUGUCAGAGUGAGAGCGAAGCGAGUUGCGAGCUCAGGCGACAAAGCGAGAGAGCAGUUUGCAACCUGCUUUAUCGCUUCAACUCGCUCAGAGCGCACGAUUGUUUUUUUGUAUCCUGU .....(((.((....))(((((((..(((..((((.(((((..((((((.(((((((........))))))).))))))....)))))..)))))))...)))))))........))). (-33.09 = -32.97 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:12 2006