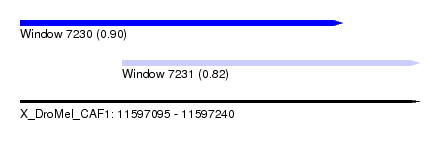
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,597,095 – 11,597,240 |
| Length | 145 |
| Max. P | 0.904525 |

| Location | 11,597,095 – 11,597,212 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.86 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -24.14 |
| Energy contribution | -25.50 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11597095 117 + 22224390 UUUUCCC-UAUUUUCAGAUCCGAUCGCCACGCCCACAGCGCCAACAUGUGGAGCCCACAAAAAGGCCCUACACGACUGUGGGACCUUAGGUUUAGUAGCUGUAUAUUUAUCUUACACU .......-......(((....((((......(((((((((......(((((.(((........))).))))))).)))))))......))))......)))................. ( -26.90) >DroVir_CAF1 34973 116 + 1 -CGCUUU-CUUUUGCAGAUAUAGCCGCAGCGCCCACAUCGCCAGCAUGUGGAGCCCACAAAAAGGUCCUACACGGCUGUGGGACCUUAGAUUUAGUAGCUGUAUAUUUAUUUUACAUU -.((...-.....))..(((((((..(.(.(((((((((....).)))))).)).).....(((((((((((....))))))))))).......)..))))))).............. ( -38.90) >DroGri_CAF1 41936 113 + 1 -CA---C-CCAUUCCAGAUAUAGGCGAAGCGCCCACAUCGCCAGCAUGUGGAGCCCACAAAAAGGUCCUACACGGCUGUGGGACCUUAGACUUAGUAGCUGUAUAUUUAUCUUACAUU -..---.-.......((((((((((((.(......).)))))(((.(((((...)))))..(((((((((((....)))))))))))..........))).))))))).......... ( -35.70) >DroWil_CAF1 59036 117 + 1 -UUCAUCUCGUUCUCAGAUCUAGACGCAGCGCCCACAUUGCCAAUAUGUGGAGUCCACAAAAAGGUCCUACACGUCUGUGGGACAUUAGAUUUAGUAGCUGUAUAUUUAUUUUACAUU -........(((((.((((((((.....(.((((((((.......)))))).)).)........((((((((....)))))))).)))))))))).)))((((.........)))).. ( -28.80) >DroMoj_CAF1 40131 116 + 1 -UCUUCU-UCUUUGCAGAUAUAGCCGCAGCGUCCACAUCUCCAGCAUGUGGAGCCCACAAAAAGGUCCUACACGGCUGUGGGACCUUAGACUUAGUAGCUGUAUAUUUAUUUUACAUU -......-.........(((((((..(.((.((((((((....).))))))))).......(((((((((((....))))))))))).......)..))))))).............. ( -37.40) >DroAna_CAF1 40364 116 + 1 -CUUUUU-UAAUUUCAGUUUCGAUUGCCACUUCCACAGAACCAAUAUGUGGAGUCCACAGAAGGGCCCUACACGACUGUGGGACCUUAGGUUUAGUAGCUGUAUAUUUAUCUUACACC -......-......(((((.(....(((..(((((((.........))))))).......((((..((((((....)))))).)))).)))...).)))))................. ( -27.60) >consensus _CUUUUC_CAUUUUCAGAUAUAGCCGCAGCGCCCACAUCGCCAACAUGUGGAGCCCACAAAAAGGUCCUACACGGCUGUGGGACCUUAGAUUUAGUAGCUGUAUAUUUAUCUUACAUU .........................(((((((((((((.......)))))).)).......(((((((((((....)))))))))))..........)))))................ (-24.14 = -25.50 + 1.36)


| Location | 11,597,132 – 11,597,240 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 83.01 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -21.54 |
| Energy contribution | -21.18 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11597132 108 + 22224390 CGCCAACAUGUGGAGCCCACAAAAAGGCCCUACACGACUGUGGGACCUUAGGUUUAGUAGCUGUAUAUUUAUCUUACACUUAAUGGUAAGUAAGCA--AAAUCAAAUUGU .(((....(((((...)))))..((((.((((((....)))))).)))).)))........(((.((((((((...........)))))))).)))--............ ( -26.40) >DroVir_CAF1 35009 104 + 1 CGCCAGCAUGUGGAGCCCACAAAAAGGUCCUACACGGCUGUGGGACCUUAGAUUUAGUAGCUGUAUAUUUAUUUUACAUUUAAUGGUGAGUACAAUUUGAAA---G---U (((((((.(((((...)))))..(((((((((((....)))))))))))..........))((((.........)))).....)))))..............---.---. ( -30.30) >DroGri_CAF1 41969 104 + 1 CGCCAGCAUGUGGAGCCCACAAAAAGGUCCUACACGGCUGUGGGACCUUAGACUUAGUAGCUGUAUAUUUAUCUUACAUUUAAUGGUGAGUAUAACUAACAU---A---U ...((((.(((((...)))))..(((((((((((....)))))))))))..........))))((((((((((...........))))))))))........---.---. ( -33.90) >DroWil_CAF1 59073 105 + 1 UGCCAAUAUGUGGAGUCCACAAAAAGGUCCUACACGUCUGUGGGACAUUAGAUUUAGUAGCUGUAUAUUUAUUUUACAUUUAACGGUAAGUCAAAU--AAAU--AAUU-A ((((....(((((...))))).....((((((((....))))))))..........((((..(((....))).)))).......))))........--....--....-. ( -21.70) >DroYak_CAF1 29933 93 + 1 CCCCAACAUGUGGAGCCCACAAAAAGGCCCUACACGACUGUGGGACCUUAGGUUUAGUAGCUGUAUAUUUAUCUUACACUUAAUGGUAAGUA-----------------U ..((....(((((...)))))..((((.((((((....)))))).)))).))............(((((((((...........))))))))-----------------) ( -22.40) >DroMoj_CAF1 40167 106 + 1 CUCCAGCAUGUGGAGCCCACAAAAAGGUCCUACACGGCUGUGGGACCUUAGACUUAGUAGCUGUAUAUUUAUUUUACAUUUAAUGGUAAGU-CACUUUGAAUCAUA---U (((((.....)))))....(((((((((((((((....))))))))))).((((((.((..((((.........)))).....)).)))))-)..)))).......---. ( -31.40) >consensus CGCCAACAUGUGGAGCCCACAAAAAGGUCCUACACGGCUGUGGGACCUUAGAUUUAGUAGCUGUAUAUUUAUCUUACAUUUAAUGGUAAGUAAAAU__AAAU___A___U .....((((((((...)))))..(((((((((((....)))))))))))............)))........(((((........))))).................... (-21.54 = -21.18 + -0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:08 2006