
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,595,673 – 11,595,858 |
| Length | 185 |
| Max. P | 0.993883 |

| Location | 11,595,673 – 11,595,786 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.43 |
| Mean single sequence MFE | -34.19 |
| Consensus MFE | -21.89 |
| Energy contribution | -22.17 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
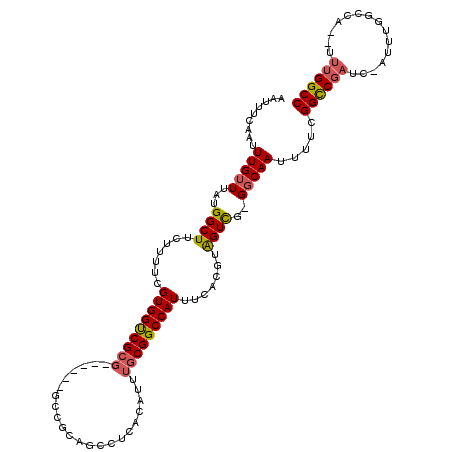
>X_DroMel_CAF1 11595673 113 + 22224390 AAUUUCAAUUUGUUUAUGGCUUCUUUUCGUGGUCGCGG--GCAGCCGCAGCCUCACAUUUGCGGCCAUUUCACGUAGUCG-GGCAAUUUUCGGCCGAUCCAUUUGGCCA--UUUGGCC .................((((.......((((((((((--((.......)))).......)))))))).......))))(-(.(((.....((((((.....)))))).--.))).)) ( -33.35) >DroSec_CAF1 27900 109 + 1 AAUUUCAAUUUGUUUAUGGCUUCUUUUCGUGGUCGCG------GCCGCAGCCUCACAUUUGCGGCCAUUUCACGUAGUCG-GGCAAUUUUCGGCCGAUCCAUUUGGCCA--UUUGGCC .................((((......(((((....(------(((((((........))))))))...))))).))))(-(.(((.....((((((.....)))))).--.))).)) ( -35.60) >DroSim_CAF1 27289 109 + 1 AAUUUCAAUUUGUUUAUGGCUUCUUUUCGUGGUCGCG------GCCGCAGCCUCACAUUUGCGGCCAUUUCACGUAGUCG-GGCAAUUUUCGGCCGAUCCAUUUGGCCA--UUUGGCC .................((((......(((((....(------(((((((........))))))))...))))).))))(-(.(((.....((((((.....)))))).--.))).)) ( -35.60) >DroEre_CAF1 27934 114 + 1 AAUUUCAAUUUGUUUAUGGCUUCUUUUCGUGGUCGCGGUCGCGGUCGCAGCCUCACAUUUGCGGCCAUUUCACGUAGUCG-GGCAAUUUUCGGCCGAUC-AUUUGGCCG--UUUGGCC .................((((......(((((..((....))((((((((........))))))))...))))).))))(-(.(((....(((((((..-..)))))))--.))).)) ( -38.60) >DroYak_CAF1 28458 105 + 1 AAUUUCAAUUUGUUUAUGGCUUCUUUUCGUGGUCGCGC--AGCUCCACAGCCUCACAUUUGCGGCCAUUUCACGUAGUUG-GGCAAUUUUCGGCCGAUC--------CA--UUUGGCC ...((((((((((..((((((.......((((..((..--.)).)))).((.........))))))))...))).)))))-))........((((((..--------..--.)))))) ( -26.40) >DroAna_CAF1 38687 96 + 1 AAUUUCAAUUUGUUUAUGGCUUCUUUUCGUGGCCG------------------CACAUUUGCGGCCAUUUGACGGGGUCUUGCCAAUUUUCGGUCAGUU----UCCCCCCCGGGGGCC .................(((((((....(((((((------------------((....))))))))).....((((.((.(((.......))).))..----.))))...))))))) ( -35.60) >consensus AAUUUCAAUUUGUUUAUGGCUUCUUUUCGUGGUCGCG______GCCGCAGCCUCACAUUUGCGGCCAUUUCACGUAGUCG_GGCAAUUUUCGGCCGAUC_AUUUGGCCA__UUUGGCC .........(((((...((((.......(((((((((......................))))))))).......))))..))))).....((((((...............)))))) (-21.89 = -22.17 + 0.28)


| Location | 11,595,711 – 11,595,826 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 87.44 |
| Mean single sequence MFE | -41.06 |
| Consensus MFE | -34.18 |
| Energy contribution | -34.74 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
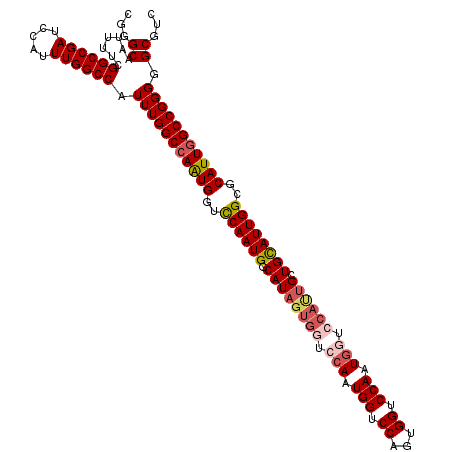
>X_DroMel_CAF1 11595711 115 + 22224390 --GCAGCCGCAGCCUCACAUUUGCGGCCAUUUCACGUAGUCGGGCAAUUUUCGGCCGAUCCAUUUGGCCAUUUGGCCAAUGGUCCAAUGGCAUACUGGUCCAAUGGUCCAGUGGUCC --((.(((((((........)))))))((((.......(((((.(.......).)))))(((((.((((....)))))))))...)))))).((((((.((...)).)))))).... ( -43.30) >DroSec_CAF1 27937 112 + 1 -----GCCGCAGCCUCACAUUUGCGGCCAUUUCACGUAGUCGGGCAAUUUUCGGCCGAUCCAUUUGGCCAUUUGGCCAGUGGUUCAAUGGCAUAGUGGUCCAAUGGUCCAGUGGUCC -----(((((((........)))))))...........(((((.(.......).)))))(((((.(((((((.(((((.((((......)).)).))))).))))))).)))))... ( -41.80) >DroSim_CAF1 27326 112 + 1 -----GCCGCAGCCUCACAUUUGCGGCCAUUUCACGUAGUCGGGCAAUUUUCGGCCGAUCCAUUUGGCCAUUUGGCCAGUGGUCCAAUGGCAUAGUGGUCCAAUGGUCCAGUGGUCC -----(((((((........)))))))...........(((((.(.......).)))))(((((.(((((((.(((((.(((((....))).)).))))).))))))).)))))... ( -43.30) >DroEre_CAF1 27972 116 + 1 UCGCGGUCGCAGCCUCACAUUUGCGGCCAUUUCACGUAGUCGGGCAAUUUUCGGCCGAUC-AUUUGGCCGUUUGGCCAAUGGUCCAAUGGCAUAGUGGUCCAGUGGUCCAUUGGUCC ..((((((((((........))))))))......((....)).))......(((((((..-..)))))))...(((((((((.(((.((((......).))).))).))))))))). ( -49.30) >DroYak_CAF1 28496 99 + 1 --AGCUCCACAGCCUCACAUUUGCGGCCAUUUCACGUAGUUGGGCAAUUUUCGGCCGAUC--------CAUUUGGCCAAUGGUUCAAU--------GGUCCAAUGGUCCAAUGGUCC --.(((....)))....((((.(.(((((((..((...(((((((....((.((((((..--------...))))))))..)))))))--------.))..)))))))))))).... ( -27.60) >consensus _____GCCGCAGCCUCACAUUUGCGGCCAUUUCACGUAGUCGGGCAAUUUUCGGCCGAUCCAUUUGGCCAUUUGGCCAAUGGUCCAAUGGCAUAGUGGUCCAAUGGUCCAGUGGUCC .....(((((((........)))))))...........(((((.(.......).)))))......(((((((.(((((.(((.(((.........))).))).))))).))))))). (-34.18 = -34.74 + 0.56)

| Location | 11,595,749 – 11,595,858 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.46 |
| Mean single sequence MFE | -51.00 |
| Consensus MFE | -45.25 |
| Energy contribution | -47.00 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
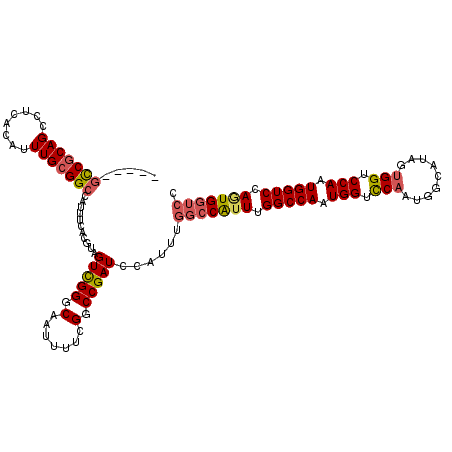
>X_DroMel_CAF1 11595749 109 + 22224390 CGGGCAAUUUUCGGCCGAUCCAUUUGGCCAUUUGGCCAAUGGUCCAAUGGCAUACUGGUCCAAUGGUCCAGUGGUCCAAU--------GCUGUAUUGGCGCAUUGGCCGGGGGCGUC ...((.......((((((.....)))))).((((((((((((.(((((((((((((((.((...)).)))))......))--------)))..)))))).))))))))))).))... ( -45.60) >DroSec_CAF1 27972 117 + 1 CGGGCAAUUUUCGGCCGAUCCAUUUGGCCAUUUGGCCAGUGGUUCAAUGGCAUAGUGGUCCAAUGGUCCAGUGGUCCAAUGAUCCAUUGCUGCAUUGGCGCAUUGGCCGGGGGCGUC ...((.......((((((.....)))))).(((((((((((((.(((((.((((((((..((.(((.((...)).))).))..)))))).))))))))).))))))))))).))... ( -49.10) >DroSim_CAF1 27361 117 + 1 CGGGCAAUUUUCGGCCGAUCCAUUUGGCCAUUUGGCCAGUGGUCCAAUGGCAUAGUGGUCCAAUGGUCCAGUGGUCCAAUGGUCCAUUGCUGCAUUGGCGCAUUGGCCGGGGGCGUC ...((.......((((((.....)))))).((((((((((((.((((((.((((((((.(((.(((.((...)).))).))).)))))).))))))))).))))))))))).))... ( -56.00) >DroEre_CAF1 28012 116 + 1 CGGGCAAUUUUCGGCCGAUC-AUUUGGCCGUUUGGCCAAUGGUCCAAUGGCAUAGUGGUCCAGUGGUCCAUUGGUCCAUUGGUGCACUGGUGCAUUGGUGCAUUCGCCGGGGGCGUC .((.(((....(((((((..-..))))))).))).))....((((..((((.((((((.((((((...)))))).))))))((((((..(....)..))))))..)))).))))... ( -53.30) >consensus CGGGCAAUUUUCGGCCGAUCCAUUUGGCCAUUUGGCCAAUGGUCCAAUGGCAUAGUGGUCCAAUGGUCCAGUGGUCCAAUGGUCCAUUGCUGCAUUGGCGCAUUGGCCGGGGGCGUC ...((.......((((((.....)))))).(((((((((((..((((((.((((((((.(((.(((.((...)).))).))).)))))).))))))))..))))))))))).))... (-45.25 = -47.00 + 1.75)


| Location | 11,595,749 – 11,595,858 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 90.46 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -32.16 |
| Energy contribution | -34.60 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
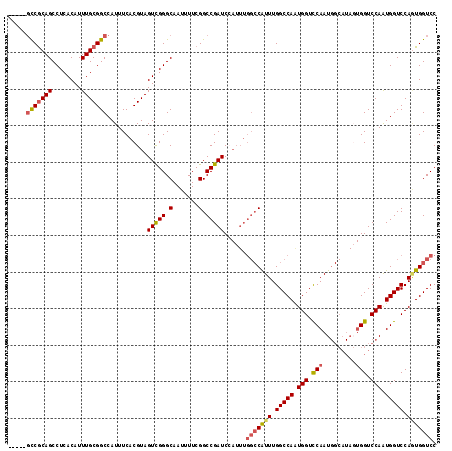
>X_DroMel_CAF1 11595749 109 - 22224390 GACGCCCCCGGCCAAUGCGCCAAUACAGC--------AUUGGACCACUGGACCAUUGGACCAGUAUGCCAUUGGACCAUUGGCCAAAUGGCCAAAUGGAUCGGCCGAAAAUUGCCCG ...(((.(((((((((((.........))--------))))).))(((((.((...)).)))))........)).(((((((((....)))).)))))...)))............. ( -40.20) >DroSec_CAF1 27972 117 - 1 GACGCCCCCGGCCAAUGCGCCAAUGCAGCAAUGGAUCAUUGGACCACUGGACCAUUGGACCACUAUGCCAUUGAACCACUGGCCAAAUGGCCAAAUGGAUCGGCCGAAAAUUGCCCG ........(((((.......(((((((....(((.(((.(((.((...)).))).))).)))...)).)))))..(((.(((((....)))))..)))...)))))........... ( -37.90) >DroSim_CAF1 27361 117 - 1 GACGCCCCCGGCCAAUGCGCCAAUGCAGCAAUGGACCAUUGGACCACUGGACCAUUGGACCACUAUGCCAUUGGACCACUGGCCAAAUGGCCAAAUGGAUCGGCCGAAAAUUGCCCG ........(((((......((((((((....(((.(((.(((.((...)).))).))).)))...)).)))))).(((.(((((....)))))..)))...)))))........... ( -43.20) >DroEre_CAF1 28012 116 - 1 GACGCCCCCGGCGAAUGCACCAAUGCACCAGUGCACCAAUGGACCAAUGGACCACUGGACCACUAUGCCAUUGGACCAUUGGCCAAACGGCCAAAU-GAUCGGCCGAAAAUUGCCCG .........(((((.(((((..........)))))....(((.(((((((.(((.(((.(......)))).))).))))))))))..(((((....-....)))))....))))).. ( -42.30) >consensus GACGCCCCCGGCCAAUGCGCCAAUGCAGCAAUGGACCAUUGGACCACUGGACCAUUGGACCACUAUGCCAUUGGACCACUGGCCAAAUGGCCAAAUGGAUCGGCCGAAAAUUGCCCG ...((....((((((((..((((((((....(((.(((.(((.((...)).))).))).)))...)).))))))..))))))))...(((((.........)))))......))... (-32.16 = -34.60 + 2.44)

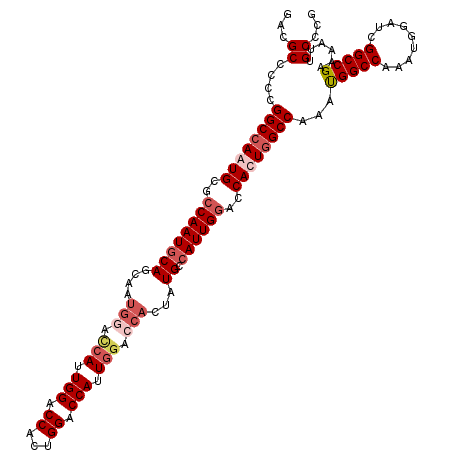
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:06 2006