
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,565,978 – 11,566,093 |
| Length | 115 |
| Max. P | 0.976098 |

| Location | 11,565,978 – 11,566,093 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.65 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -12.67 |
| Energy contribution | -13.73 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
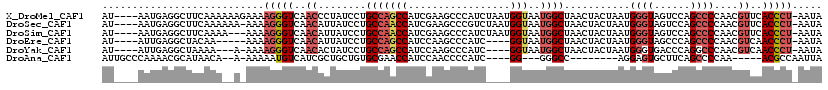
>X_DroMel_CAF1 11565978 115 + 22224390 AU----AAUGAGGCUUCAAAAAAGAAAAGGGUCAACCCUAUCCUGCCAGCCAUCGAAGCCCAUCUAAUGGUAAUGGCUAACUACUAAUGGGUAGUCCAGCCCCAACGUUCACCCU-AAUA ..----.(((.((((((..........((((....)))).....(....)....)))))))))....(((....((((.((((((....))))))..)))))))...........-.... ( -28.10) >DroSec_CAF1 7591 114 + 1 AU----AAUGAGGCUUCAAAAAA-AAAAGGGUCAACAUUAUCCUGCCAACCAUCGAAGCCCGUCUAAUGGUAAUGGCUAACUACUAAUGGGUAGUCCAGCCCCAACGUUCACCCU-AAUA ..----.................-...(((((.(((.......(((((.....((.....)).....)))))..((((.((((((....))))))..)))).....))).)))))-.... ( -26.60) >DroSim_CAF1 8403 112 + 1 AU----AAUGAGGCUUCAAAA---AAAAGGGUCAACAUUAUCCUGCCAACCAUCGAAGCCCAUCUAAUGGUAAUGGCUAACUACUAAUGGGUAGUCCAGCCCCAACGUUCACCCU-AAUA ..----.(((.((((((....---...(((((.......)))))..........)))))))))....(((....((((.((((((....))))))..)))))))...........-.... ( -25.83) >DroEre_CAF1 7155 106 + 1 AU----AUUGAGGCUACAA-----AAAAGGGUCAACAUUAUCCUGCCAGCCAUCCAAGCCCAUC----GGUAAUGGCUAACUACUAAUGGGUAGCCCAGCCCCAACGUCAACCCU-AAUA ..----.(((.((((((..-----...(((((.......)))))...((((((....(((....----))).))))))............)))))))))................-.... ( -23.70) >DroYak_CAF1 7654 107 + 1 AU----AUUGAGGCUAAAA---A-AAAAGGGUCAACACUAUCCUGCCAGCCAUCCAAGCCCAUC----GGUAAUGGCUAACUACUAAUGGGUGACCCAGGCCCAACGUCAACCCU-AAUA .(----((((.((......---.-....((((((...((((...(..((((((....(((....----))).))))))..).....)))).)))))).(((.....)))..)).)-)))) ( -25.20) >DroAna_CAF1 8913 97 + 1 AUUGCCCAAAACGCAUAACA--A-AAAAAUGUCAUCGCUGCUGUGCGAACCAUCCAACCCCAUC----GG---GGGCC--------AGGAGUGCUUCAGCCCCAA-----ACGCCAAUUA ...((.......((((.(((--.-.....))).)).)).((((.(((.....(((..((((...----.)---)))..--------.))).)))..)))).....-----..))...... ( -20.40) >consensus AU____AAUGAGGCUUCAAA__A_AAAAGGGUCAACACUAUCCUGCCAACCAUCCAAGCCCAUC____GGUAAUGGCUAACUACUAAUGGGUAGUCCAGCCCCAACGUUCACCCU_AAUA ...........................(((((..((........(((((((.................)))..))))...........((((......))))....))..)))))..... (-12.67 = -13.73 + 1.06)

| Location | 11,565,978 – 11,566,093 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.65 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -23.31 |
| Energy contribution | -24.12 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
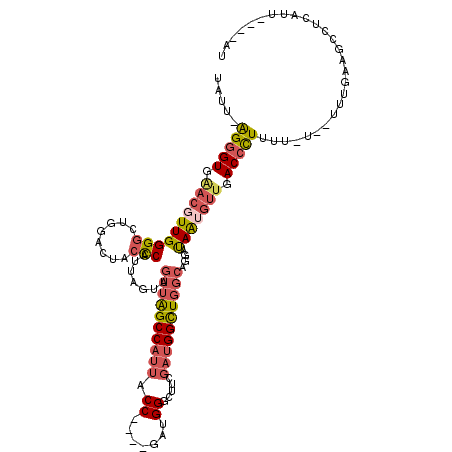
>X_DroMel_CAF1 11565978 115 - 22224390 UAUU-AGGGUGAACGUUGGGGCUGGACUACCCAUUAGUAGUUAGCCAUUACCAUUAGAUGGGCUUCGAUGGCUGGCAGGAUAGGGUUGACCCUUUUCUUUUUUGAAGCCUCAUU----AU ....-..(((((.(....)(((((.(((((......)))))))))).)))))....((((((((((((..(.....((((.((((....)))).)))))..)))))))).))))----.. ( -37.70) >DroSec_CAF1 7591 114 - 1 UAUU-AGGGUGAACGUUGGGGCUGGACUACCCAUUAGUAGUUAGCCAUUACCAUUAGACGGGCUUCGAUGGUUGGCAGGAUAAUGUUGACCCUUUU-UUUUUUGAAGCCUCAUU----AU ....-..(((((.(((((((((((.(((((......))))))))))....)))....)))((((((((.(((..(((......)))..))).....-....)))))))))))))----.. ( -36.30) >DroSim_CAF1 8403 112 - 1 UAUU-AGGGUGAACGUUGGGGCUGGACUACCCAUUAGUAGUUAGCCAUUACCAUUAGAUGGGCUUCGAUGGUUGGCAGGAUAAUGUUGACCCUUUU---UUUUGAAGCCUCAUU----AU ....-..(((((.(....)(((((.(((((......)))))))))).)))))....((((((((((((.(((..(((......)))..))).....---..)))))))).))))----.. ( -38.40) >DroEre_CAF1 7155 106 - 1 UAUU-AGGGUUGACGUUGGGGCUGGGCUACCCAUUAGUAGUUAGCCAUUACC----GAUGGGCUUGGAUGGCUGGCAGGAUAAUGUUGACCCUUUU-----UUGUAGCCUCAAU----AU ....-(((((..(((((((((........))).......((((((((((.((----....))....))))))))))....))))))..)))))...-----.............----.. ( -35.90) >DroYak_CAF1 7654 107 - 1 UAUU-AGGGUUGACGUUGGGCCUGGGUCACCCAUUAGUAGUUAGCCAUUACC----GAUGGGCUUGGAUGGCUGGCAGGAUAGUGUUGACCCUUUU-U---UUUUAGCCUCAAU----AU ....-(((((..((((((..((((((...))).......((((((((((.((----....))....))))))))))))).))))))..)))))...-.---.............----.. ( -35.80) >DroAna_CAF1 8913 97 - 1 UAAUUGGCGU-----UUGGGGCUGAAGCACUCCU--------GGCCC---CC----GAUGGGGUUGGAUGGUUCGCACAGCAGCGAUGACAUUUUU-U--UGUUAUGCGUUUUGGGCAAU ........((-----(..((((.(((.((.(((.--------(((((---(.----...))))))))))).)))........((.((((((.....-.--))))))))))))..)))... ( -32.10) >consensus UAUU_AGGGUGAACGUUGGGGCUGGACUACCCAUUAGUAGUUAGCCAUUACC____GAUGGGCUUCGAUGGCUGGCAGGAUAAUGUUGACCCUUUU_U__UUUGAAGCCUCAUU____AU .....(((((.((((((((((........))).......((((((((((.((.......)).....))))))))))....))))))).)))))........................... (-23.31 = -24.12 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:55 2006