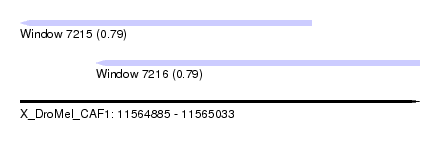
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,564,885 – 11,565,033 |
| Length | 148 |
| Max. P | 0.790984 |

| Location | 11,564,885 – 11,564,993 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.35 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
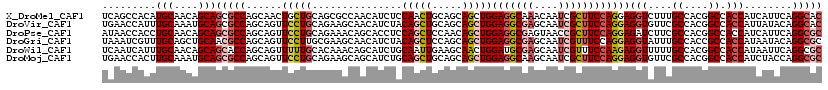
>X_DroMel_CAF1 11564885 108 - 22224390 CCAACAUCUCCAACUGCAGCAGCUGGAGGCAAACAAUCGCUUCCAGGAGGUCUUUGCCACGGCCACCAUCAUUCAGGCACAUCCGCAUCCCCAUCCACAUCCCAGGGA .....((((((....((....))(((((((........)))))))))))))...(((....(((...........)))......)))((((.............)))) ( -26.62) >DroVir_CAF1 9973 108 - 1 GCAACAUCUACAGCUGCAGCAGCUGGAGGCGAGCAAUCGCUUCCAGGAGGUGUUCGCCACGGCCACCAUUAUACAGGCACAUCCACAUCCGCAUCCGCACUUGCGUGA (((((((((...((....))..((((((((((....)))))))))).))))))).))(((((((...........)))............((....)).....)))). ( -39.00) >DroPse_CAF1 8245 102 - 1 ACAGCACCUCCAGCUCCAACAGCUGGAGGCGAGUAACCGCUUCCAGGAGAUCUUCGCCACGGCCACCAUCAUUCAGGCGCAUCCGCAUCCCAA------UCCCCGGGA ...((..((((((((.....))))(((((((......))))))).)))).....((((.................)))).....)).((((..------.....)))) ( -32.23) >DroGri_CAF1 10164 96 - 1 GCAACAUCUACAGCUCCAGCAGCUGGAGGCGAGCAAUCGUUUCCAGGAGGUAUUUGCCACCGCCACCAUAAUACAGGCGCAUCCGCAU------------UCACGCGA ((((.((((...((....))..((((((((((....)))))))))).))))..))))...((((...........))))....(((..------------....))). ( -30.70) >DroWil_CAF1 29929 96 - 1 ACAGCAUCUGCAAUUGAAGCAACUGGAUGCGAGCAAUCGUUUCCAAGAGGUUUUUGCCACGGCCACCAUAAUUCAGGCGCAUCCCCA------GUCGCUU------GA ...((....(((...(((((..(((((.((((....)))).))).))..))))))))....)).........((((((((.......------).)))))------)) ( -26.40) >DroMoj_CAF1 9994 96 - 1 GCAGCAUCUGCAGCUGCAGCAGCUGGAGGCAAGCAAUCGCUUCCAGGAGGUGUUCGCCACGGCCACCAUCUACCAGGCGCAUCAGCAA------------CCACGUGA (((((((((.((((((...)))))((((((........)))))).).))))))).))(((((((...........)))((....))..------------...)))). ( -34.80) >consensus GCAACAUCUACAGCUGCAGCAGCUGGAGGCGAGCAAUCGCUUCCAGGAGGUCUUCGCCACGGCCACCAUCAUUCAGGCGCAUCCGCAU___________UCCACGGGA ......................((((((((((....))))))))))..(((....)))...(((...........))).............................. (-19.12 = -19.90 + 0.78)



| Location | 11,564,913 – 11,565,033 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.89 |
| Mean single sequence MFE | -41.26 |
| Consensus MFE | -25.99 |
| Energy contribution | -27.27 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11564913 120 - 22224390 UCAGCCACAUGCAACAGCAGCGCCAGCAACUGCUGCAGCGCCAACAUCUCCAACUGCAGCAGCUGGAGGCAAACAAUCGCUUCCAGGAGGUCUUUGCCACGGCCACCAUCAUUCAGGCAC ...(((...(((....)))(.(((.(((((((((((((...............)))))))))((((((((........)))))))).......))))...))))...........))).. ( -43.16) >DroVir_CAF1 10001 120 - 1 UGAACCAUUUGCAAAUGCAGCGCCAGCAGUUCCUGCAGAAGCAACAUCUACAGCUGCAGCAGCUGGAGGCGAGCAAUCGCUUCCAGGAGGUGUUCGCCACGGCCACCAUUAUACAGGCAC ....((..((((...((((((....((((...))))(((.......)))...))))))))))((((((((((....))))))))))..((((..((...))..))))........))... ( -44.30) >DroPse_CAF1 8267 120 - 1 AUAACCACCUGCAACAGCAGCGCCAGCAGUUCCUGCAGAAACAGCACCUCCAGCUCCAACAGCUGGAGGCGAGUAACCGCUUCCAGGAGAUCUUCGCCACGGCCACCAUCAUUCAGGCGC .........(((....)))(((((.((((...)))).(((.......((((((((.....))))(((((((......))))))).))))......((....))........))).))))) ( -39.70) >DroGri_CAF1 10180 120 - 1 UAAAUCGUUUGCAGCUGCAACGCCAGCAGUUCCUUGCGAAGCAACAUCUACAGCUCCAGCAGCUGGAGGCGAGCAAUCGUUUCCAGGAGGUAUUUGCCACCGCCACCAUAAUACAGGCGC ....((((..(.((((((.......)))))).)..)))).((((.((((...((....))..((((((((((....)))))))))).))))..))))...((((...........)))). ( -40.40) >DroWil_CAF1 29945 120 - 1 UCAAUCAUUUGCAACAGCAGCACCAGCAGUUUUUGCACAAACAGCAUCUGCAAUUGAAGCAACUGGAUGCGAGCAAUCGUUUCCAAGAGGUUUUUGCCACGGCCACCAUAAUUCAGGCGC .........(((....)))((.((.((((....(((.......))).))))...((((.....((((.((((....)))).))))...(((....((....)).)))....)))))).)) ( -31.30) >DroMoj_CAF1 10010 120 - 1 UGAACCACUUGCAAAUGCAGCGCCAGCAGUUCCUGCAGAAGCAGCAUCUGCAGCUGCAGCAGCUGGAGGCAAGCAAUCGCUUCCAGGAGGUGUUCGCCACGGCCACCAUCUACCAGGCGC .........(((....)))(((((...((((.((((((..((((...))))..)))))).))))((((((........)))))).(((((((...((....))...))))).)).))))) ( -48.70) >consensus UCAACCACUUGCAACAGCAGCGCCAGCAGUUCCUGCAGAAGCAACAUCUACAGCUGCAGCAGCUGGAGGCGAGCAAUCGCUUCCAGGAGGUCUUCGCCACGGCCACCAUCAUUCAGGCGC .........(((....)))(((((......(((((...............(((((.....)))))(((((((....))))))))))))(((....((....)).)))........))))) (-25.99 = -27.27 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:53 2006