| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,537,319 – 11,537,409 |
| Length | 90 |
| Max. P | 0.991318 |

| Location | 11,537,319 – 11,537,409 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 88.76 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -22.75 |
| Energy contribution | -22.75 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11537319 90 + 22224390 CGAUAAUAAAAUCAAGCAAUCGGGCCGGCAGACCCAACUCAUGACCAUAU------GC-AACCCUGCCGGCCCAGCAUUCGUGUGUCUGUUUUCCGU- .(((......)))(((((...((((((((((........((((....)))------).-....)))))))))).((((....)))).))))).....- ( -26.22) >DroSec_CAF1 3112 90 + 1 CGAUAAUAAAAUCAAACAAUCGGGCCGGCAGACCCAACUAAUGACCAUAU------GC-AACCCUGCCGGCCCAGCAUUCGUGUGUCUGUGUUUCGU- .(((......)))(((((...((((((((((...................------..-....)))))))))).((((....))))...)))))...- ( -26.21) >DroSim_CAF1 3147 90 + 1 CGAUAAUAAAAUCAAACUAUCGGGCCGGCAGACCCAACUCAUGACCAUAU------GC-AACCCUGCCGGCCCAGCAUUCGUGUGUCUGUGUUUCGU- (((.((((.............((((((((((........((((....)))------).-....)))))))))).((((....))))...))))))).- ( -25.52) >DroEre_CAF1 3237 84 + 1 CGAUA-----AUCAAACAAUCGGGCCGGCAGACCCAACUCAUGACCAUAU------GC-AACCCUGCCGGCCCAGCAUUCGUG--UCUUUGUUCCGUU .....-----....(((((..((((((((((........((((....)))------).-....)))))))))).(((....))--)..)))))..... ( -23.92) >DroYak_CAF1 3153 90 + 1 CGAUA-----AUCAAACAAUCGGGCCGGCAGACCCAACUCAUGACCAUAUAUGUAUGCAAAACCUGCCGGCCCAGCAUUCGUG--UCUGUGUUCCGU- .....-----...........((((((((((...((.....))..((((....))))......))))))))))(((((.....--...)))))....- ( -25.50) >consensus CGAUAAUAAAAUCAAACAAUCGGGCCGGCAGACCCAACUCAUGACCAUAU______GC_AACCCUGCCGGCCCAGCAUUCGUGUGUCUGUGUUCCGU_ .(((......)))........((((((((((................................))))))))))......................... (-22.75 = -22.75 + 0.00)


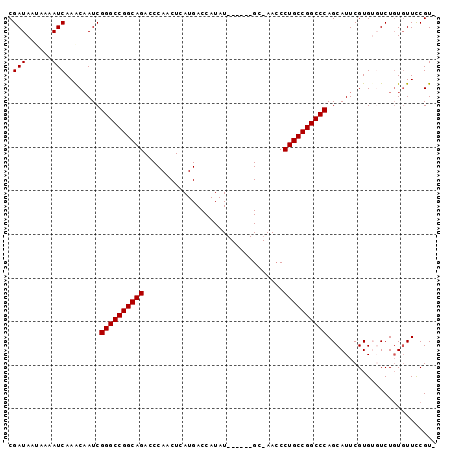
| Location | 11,537,319 – 11,537,409 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 88.76 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -26.41 |
| Energy contribution | -26.61 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.91 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11537319 90 - 22224390 -ACGGAAAACAGACACACGAAUGCUGGGCCGGCAGGGUU-GC------AUAUGGUCAUGAGUUGGGUCUGCCGGCCCGAUUGCUUGAUUUUAUUAUCG -.....................((((((((((((((.((-.(------((......)))....)).))))))))))))...))..(((......))). ( -27.10) >DroSec_CAF1 3112 90 - 1 -ACGAAACACAGACACACGAAUGCUGGGCCGGCAGGGUU-GC------AUAUGGUCAUUAGUUGGGUCUGCCGGCCCGAUUGUUUGAUUUUAUUAUCG -........((((((((....)).((((((((((((.((-..------.((.......))...)).))))))))))))..))))))............ ( -28.00) >DroSim_CAF1 3147 90 - 1 -ACGAAACACAGACACACGAAUGCUGGGCCGGCAGGGUU-GC------AUAUGGUCAUGAGUUGGGUCUGCCGGCCCGAUAGUUUGAUUUUAUUAUCG -........................(((((((((((.((-.(------((......)))....)).)))))))))))((((((........)))))). ( -29.90) >DroEre_CAF1 3237 84 - 1 AACGGAACAAAGA--CACGAAUGCUGGGCCGGCAGGGUU-GC------AUAUGGUCAUGAGUUGGGUCUGCCGGCCCGAUUGUUUGAU-----UAUCG ....((((((((.--((....))))(((((((((((.((-.(------((......)))....)).)))))))))))..))))))...-----..... ( -28.70) >DroYak_CAF1 3153 90 - 1 -ACGGAACACAGA--CACGAAUGCUGGGCCGGCAGGUUUUGCAUACAUAUAUGGUCAUGAGUUGGGUCUGCCGGCCCGAUUGUUUGAU-----UAUCG -........((((--((.......((((((((((((..(..(((((.......)).)))....)..))))))))))))..))))))..-----..... ( -30.70) >consensus _ACGGAACACAGACACACGAAUGCUGGGCCGGCAGGGUU_GC______AUAUGGUCAUGAGUUGGGUCUGCCGGCCCGAUUGUUUGAUUUUAUUAUCG .................(((((..((((((((((((.((.((..................)).)).))))))))))))...)))))............ (-26.41 = -26.61 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:43 2006