
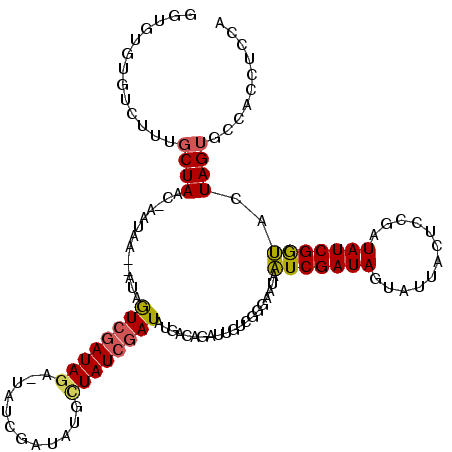
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,532,806 – 11,532,919 |
| Length | 113 |
| Max. P | 0.994492 |

| Location | 11,532,806 – 11,532,919 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.00 |
| Mean single sequence MFE | -35.34 |
| Consensus MFE | -13.12 |
| Energy contribution | -12.84 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.37 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11532806 113 + 22224390 GGGGUGUGUCUUAGCUAACAAUUACGU---GUCGAUAGA-UAUA---AUGCUAUCGAUAUGACAGAGUGUCGGGAACAAUCGAUAGCAUUACUUCGAUAUCGGUACUAGUGCCACCUCCA ((((((.((....((((........((---..(((((((-..((---(((((((((((..(((.....)))(....).)))))))))))))..))..)))))..)))))))))))).)). ( -39.80) >DroSec_CAF1 14017 118 + 1 GAUGUAUGUCUUUGCUAAA-AUUAAACAUCGUAAAUAGA-UAUCGGUAUGUUAUCGAUAUGACAGAUUGUCGGGAAUAAUCGAUAGUAUUACUCCGAUAUCGGUACUAGUGCCACCUCCA (((((.((..(((....))-).)).))))).......((-((((((.(((((((((((.((((.....))))......)))))))))))....))))))))((((....))))....... ( -30.50) >DroSim_CAF1 16529 118 + 1 GGUGUGUGUCUUUGCUAAA-CAUAGACAUCGUCGAUAGA-UAUCGAUAUGCUAUCGAUAUGACAUUUUGUCGGGAAUAAUCGAUAGUAUUACUCCGAUAUCGGUACUAGUGCCACCUCCA (((..((((((.((.....-)).))))))........((-(((((..(((((((((((.((((.....))))......))))))))))).....)))))))((((....))))))).... ( -36.50) >DroEre_CAF1 14174 115 + 1 GGCACGAGUCUUUGCUAAC-GAUGA--AUAAUCGAUAUGCC-UCGAUAAGUUAUCGAUGUCGUUUAGCUUCGUGAAUACUCGAUAGUAUCGUGCGGAUAUCGAGGCUAG-GUCACCUCUA ((((........))))...-(((..--...)))(((..(((-((((((.((((.((....))..))))((((((.(((((....)))))..)))))))))))))))...-)))....... ( -31.00) >DroYak_CAF1 14690 117 + 1 GGAGUGUGUCUUUGCUAAC-AAUAC--AUAAUCGAUAGGCUAUCGACAAGCUAUCGAUGUCGUCGGUUUUCGUGAAUACUCGAUAGUAUCGCGUUGGUAUCGAGGCUAGUGCCACCUCCA ((((.((.((..(((((((-.....--..(((((((.(((.(((((.......)))))))))))))))...((((.((((....)))))))))))))))..))(((....))))))))). ( -38.90) >consensus GGUGUGUGUCUUUGCUAAC_AAUAA__AUAGUCGAUAGA_UAUCGAUAUGCUAUCGAUAUGACAGAUUGUCGGGAAUAAUCGAUAGUAUUACUCCGAUAUCGGUACUAGUGCCACCUCCA .............((((.............((((((((............))))))))....................(((((((............)))))))..)))).......... (-13.12 = -12.84 + -0.28)


| Location | 11,532,806 – 11,532,919 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.00 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -7.40 |
| Energy contribution | -7.84 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.25 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11532806 113 - 22224390 UGGAGGUGGCACUAGUACCGAUAUCGAAGUAAUGCUAUCGAUUGUUCCCGACACUCUGUCAUAUCGAUAGCAU---UAUA-UCUAUCGAC---ACGUAAUUGUUAGCUAAGACACACCCC .((.((((......((..(((((..((.((((((((((((((.......(((.....)))..)))))))))))---))).-)))))))..---)).....((((......)))))))))) ( -34.30) >DroSec_CAF1 14017 118 - 1 UGGAGGUGGCACUAGUACCGAUAUCGGAGUAAUACUAUCGAUUAUUCCCGACAAUCUGUCAUAUCGAUAACAUACCGAUA-UCUAUUUACGAUGUUUAAU-UUUAGCAAAGACAUACAUC ....((((.(....)))))((((((((.((.....(((((((.......(((.....)))..)))))))..)).))))))-)).......(((((....(-(((....))))...))))) ( -22.70) >DroSim_CAF1 16529 118 - 1 UGGAGGUGGCACUAGUACCGAUAUCGGAGUAAUACUAUCGAUUAUUCCCGACAAAAUGUCAUAUCGAUAGCAUAUCGAUA-UCUAUCGACGAUGUCUAUG-UUUAGCAAAGACACACACC ....((((..........(((((..((((((((.......)))))))).(((.....)))(((((((((...))))))))-).)))))..(.(((((.((-.....)).)))))).)))) ( -31.40) >DroEre_CAF1 14174 115 - 1 UAGAGGUGAC-CUAGCCUCGAUAUCCGCACGAUACUAUCGAGUAUUCACGAAGCUAAACGACAUCGAUAACUUAUCGA-GGCAUAUCGAUUAU--UCAUC-GUUAGCAAAGACUCGUGCC ..(((((...-...))))).......((((((((((....))))........(((((.(((..((((((...))))))-........((....--)).))-))))))......)))))). ( -30.20) >DroYak_CAF1 14690 117 - 1 UGGAGGUGGCACUAGCCUCGAUACCAACGCGAUACUAUCGAGUAUUCACGAAAACCGACGACAUCGAUAGCUUGUCGAUAGCCUAUCGAUUAU--GUAUU-GUUAGCAAAGACACACUCC .(((((((......((..((((((.....((((((((((((.......((.....)).((....))........)))))))..))))).....--)))))-)...)).....))).)))) ( -29.20) >consensus UGGAGGUGGCACUAGUACCGAUAUCGGAGUAAUACUAUCGAUUAUUCCCGACAAUCUGUCAUAUCGAUAGCAUAUCGAUA_UCUAUCGACGAU__GUAUU_GUUAGCAAAGACACACACC ....((((..........(((((............))))).......................(((((((............)))))))...........................)))) ( -7.40 = -7.84 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:40 2006