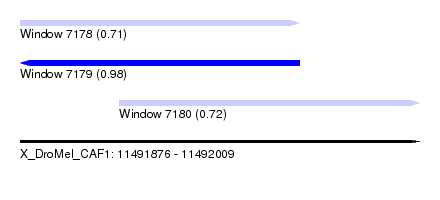
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,491,876 – 11,492,009 |
| Length | 133 |
| Max. P | 0.982981 |

| Location | 11,491,876 – 11,491,969 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -13.75 |
| Energy contribution | -13.87 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11491876 93 + 22224390 AUCUGCCACGAUUUGG---CCAACUGGGCUAAGAAGCGAUUCUCAUCUCUCUCACUUUAAAUCCUUUAAAAAGGAUUGA-GUUCAGCCAGCUGAAUU .........((((..(---(...(((((((..((((.(((....))).)).))......(((((((....))))))).)-))))))...))..)))) ( -22.20) >DroSec_CAF1 26492 90 + 1 AUCUGCCAGGAUUUGG---CCAACUGGGCUAAGAAGCGAUUCUCCACUCACUCACUUUAAAUCCUU---AAAGGAUUGA-GUUCAGCCAGCUGAAUU ........(((.((((---((.....))))))(((....))))))(((((.((.((((((....))---)))))).)))-))((((....))))... ( -24.20) >DroAna_CAF1 35391 82 + 1 --UUGUCAGGAUUUUGCUGCAAACUGGGCUAAGAUGCGAUUG----------GACUUAAUGUCCUU---UAAGGAUCGAGGUUCAGCCAACUGUAUU --(((.(((.......)))))).(((((((..(((.(....(----------(((.....))))..---...).)))..)))))))........... ( -21.00) >consensus AUCUGCCAGGAUUUGG___CCAACUGGGCUAAGAAGCGAUUCUC__CUC_CUCACUUUAAAUCCUU___AAAGGAUUGA_GUUCAGCCAGCUGAAUU .....((((...((((...))))))))((......))......................(((((((....)))))))...((((((....)))))). (-13.75 = -13.87 + 0.12)



| Location | 11,491,876 – 11,491,969 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -15.02 |
| Energy contribution | -15.47 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11491876 93 - 22224390 AAUUCAGCUGGCUGAAC-UCAAUCCUUUUUAAAGGAUUUAAAGUGAGAGAGAUGAGAAUCGCUUCUUAGCCCAGUUGG---CCAAAUCGUGGCAGAU ....(((((((((((..-..(((((((....)))))))..((((((............)))))).)))).)))))))(---(((.....)))).... ( -28.00) >DroSec_CAF1 26492 90 - 1 AAUUCAGCUGGCUGAAC-UCAAUCCUUU---AAGGAUUUAAAGUGAGUGAGUGGAGAAUCGCUUCUUAGCCCAGUUGG---CCAAAUCCUGGCAGAU ....(((((((((((((-(((.((((((---((....)))))).)).)))))((((.....)))))))).)))))))(---(((.....)))).... ( -30.90) >DroAna_CAF1 35391 82 - 1 AAUACAGUUGGCUGAACCUCGAUCCUUA---AAGGACAUUAAGUC----------CAAUCGCAUCUUAGCCCAGUUUGCAGCAAAAUCCUGACAA-- .........((((((....((((.....---..((((.....)))----------).))))....))))))(((..(........)..)))....-- ( -14.20) >consensus AAUUCAGCUGGCUGAAC_UCAAUCCUUU___AAGGAUUUAAAGUGAG_GAG__GAGAAUCGCUUCUUAGCCCAGUUGG___CCAAAUCCUGGCAGAU ......(((((((((.....(((((((....)))))))....((((............))))...))))))...((((...)))).....))).... (-15.02 = -15.47 + 0.45)


| Location | 11,491,909 – 11,492,009 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.55 |
| Mean single sequence MFE | -25.79 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.24 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11491909 100 + 22224390 ---------------CGAUUC---UCAUCUCUCUCACUUUAAAUCCUUUAAAAAGGAUUGA--GUUCAGCCAGCUGAAUUGGGGCUAAGGCCUUGCCACGAUUCGCUGCACAUGCUAAAA ---------------.((..(---(((...............((((((....)))))))))--).))(((((((.((((((.(((.........))).)))))))))).....))).... ( -24.36) >DroPse_CAF1 38618 112 + 1 AUGUUUCUGUUGCCUCUCUCUCCCCCCCUUCUCUCUCUUUAAAUCCUU---UAAGGAUUGAGAGCUCAUUCAGCUGGAUUGGGGCUAAGGCCUUGCCACGUUUCGUUGCACAC-----AG ......((((.((((......((((.((.((((..(((((((.....)---))))))..))))(((.....))).))...))))...))))..(((.(((...))).))).))-----)) ( -28.20) >DroSec_CAF1 26525 97 + 1 ---------------CGAUUC---UCCACUCACUCACUUUAAAUCCUU---AAAGGAUUGA--GUUCAGCCAGCUGAAUUGGGGCUAAGGCCUCGCCACGUUUCGCUGCACAUGCUAAAA ---------------......---...(((((.((.((((((....))---)))))).)))--))..(((((((.((((((((((....))))))....)))).)))).....))).... ( -25.30) >DroAna_CAF1 35425 79 + 1 ---------------CGAUUG-------------GACUUAAUGUCCUU---UAAGGAUCGAG-GUUCAGCCAACUGUAUUGGGGCUAAGGCCUUGCCACAAUUCGUUGCAC--------- ---------------....((-------------(((((...((((..---...))))..))-)))))((.(((...((((.(((.........))).))))..)))))..--------- ( -21.30) >DroPer_CAF1 40812 109 + 1 AUGUUUCUGUUGCCUCUCUUC---CCCCCUCUCGCUCUUUAAAUCCUU---UAAGGAUUGAGAGCUCAUUCAGCUGGAUUGGGGCUAAGGCCUUGCCACGUUUCGUUGCACAC-----AG ......((((.((((.....(---(((......((((((..(((((..---...)))))))))))..((((....)))).))))...))))..(((.(((...))).))).))-----)) ( -29.80) >consensus _______________CGAUUC___CCCCCUCUCUCACUUUAAAUCCUU___UAAGGAUUGAG_GUUCAGCCAGCUGAAUUGGGGCUAAGGCCUUGCCACGUUUCGUUGCACAC_____AA .........................................(((((((....)))))))...........((((.((((((((((....)))...))).)))).))))............ (-15.32 = -15.24 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:20 2006