
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,487,787 – 11,487,927 |
| Length | 140 |
| Max. P | 0.922317 |

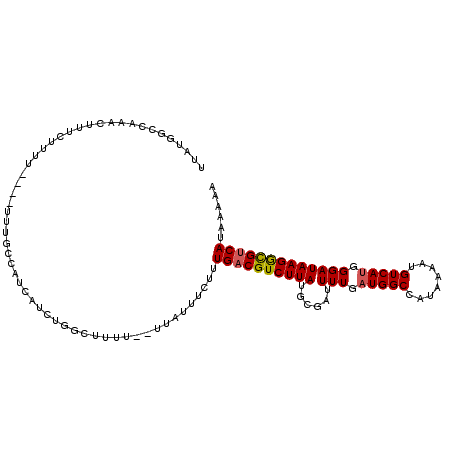
| Location | 11,487,787 – 11,487,892 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.17 |
| Mean single sequence MFE | -25.11 |
| Consensus MFE | -13.67 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
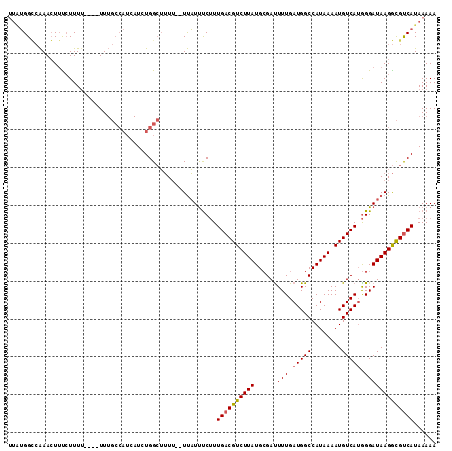
>X_DroMel_CAF1 11487787 105 + 22224390 UUAUGGGCAAACUUCCUUU-----UUUGCCAUCAUCUGGCUUU---UUAUUUCUUUGACGUCUUAUGCGAUUUUGAUGGCCAUAAAAUGUCAUGGGAUAAGGCGUCAUAAAAA .....((((((........-----))))))......((((.((---((((..(..((((((.((((((.((....)).).))))).)))))).)..)))))).))))...... ( -27.80) >DroSec_CAF1 22694 113 + 1 UUUUGGCCAAACUUUCUUUUUUUUUUUGCCAUCAUCUGGCUUUUUUUUAUUUCUUUGACGUCUUAUGCGACUUUGAUGGCCAUAAAAUGUCAUGGGAUAAGGCGUCAUAAAAA .....(((...................((((.....))))......((((..(..((((((.(((((.(.((.....)))))))).)))))).)..))))))).......... ( -25.30) >DroAna_CAF1 31116 99 + 1 AAGGGGUGGAAAUGUAUUUU----UU--------UCUGGGGGUU--UCUUCCCAUUGACGUCUUAUGAGAUUUUGAUGGCCAUAAAAUGUCAAAGGAUAAGAUGCCAUAAAAA .............(((((((----.(--------(((.((((..--...)))).(((((((.(((((.............))))).))))))))))).)))))))........ ( -22.22) >consensus UUAUGGCCAAACUUUCUUUU____UUUGCCAUCAUCUGGCUUUU__UUAUUUCUUUGACGUCUUAUGCGAUUUUGAUGGCCAUAAAAUGUCAUGGGAUAAGGCGUCAUAAAAA .......................................................((((((((((......(((.(((((........))))).)))))))))))))...... (-13.67 = -13.90 + 0.23)

| Location | 11,487,787 – 11,487,892 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 74.17 |
| Mean single sequence MFE | -19.93 |
| Consensus MFE | -10.59 |
| Energy contribution | -12.04 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11487787 105 - 22224390 UUUUUAUGACGCCUUAUCCCAUGACAUUUUAUGGCCAUCAAAAUCGCAUAAGACGUCAAAGAAAUAA---AAAGCCAGAUGAUGGCAAA-----AAAGGAAGUUUGCCCAUAA (((((.(((((.((((((((((((....))))))...........).))))).))))).)))))...---...((((.....))))...-----...((........)).... ( -20.50) >DroSec_CAF1 22694 113 - 1 UUUUUAUGACGCCUUAUCCCAUGACAUUUUAUGGCCAUCAAAGUCGCAUAAGACGUCAAAGAAAUAAAAAAAAGCCAGAUGAUGGCAAAAAAAAAAAGAAAGUUUGGCCAAAA ...(((((...........))))).......((((((.....(((......)))...................((((.....))))..................))))))... ( -20.60) >DroAna_CAF1 31116 99 - 1 UUUUUAUGGCAUCUUAUCCUUUGACAUUUUAUGGCCAUCAAAAUCUCAUAAGACGUCAAUGGGAAGA--AACCCCCAGA--------AA----AAAAUACAUUUCCACCCCUU (((((.(((..((((.(((.(((((.((((((((...........)))))))).))))).)))))))--.....))).)--------))----)).................. ( -18.70) >consensus UUUUUAUGACGCCUUAUCCCAUGACAUUUUAUGGCCAUCAAAAUCGCAUAAGACGUCAAAGAAAUAA__AAAAGCCAGAUGAUGGCAAA____AAAAGAAAGUUUGACCAAAA (((((.(((((.(((((.((((((....)))))).............))))).))))).))))).........((((.....))))........................... (-10.59 = -12.04 + 1.45)



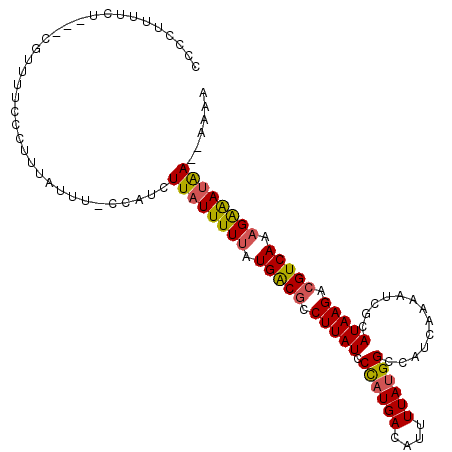
| Location | 11,487,822 – 11,487,927 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.32 |
| Mean single sequence MFE | -16.97 |
| Consensus MFE | -12.04 |
| Energy contribution | -12.27 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
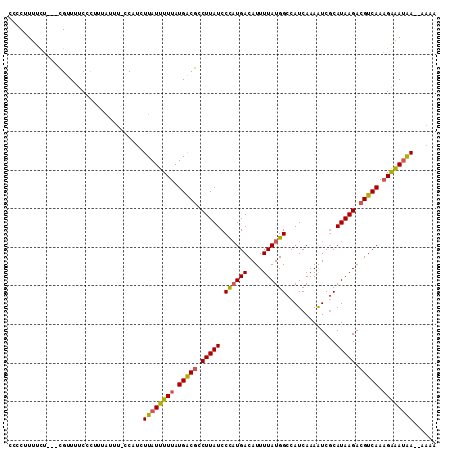
>X_DroMel_CAF1 11487822 105 - 22224390 CCCCUUUUCU---CGUUUUCCCUUUAUUUUCCAGCUUAUUUUUAUGACGCCUUAUCCCAUGACAUUUUAUGGCCAUCAAAAUCGCAUAAGACGUCAAAGAAAUAA---AAA ..........---......................((((((((.(((((.((((((((((((....))))))...........).))))).))))).))))))))---... ( -16.80) >DroSec_CAF1 22734 108 - 1 CCCCUUUUCU---CGUUUUCCCUUUUUUCCGCAUCUUAUUUUUAUGACGCCUUAUCCCAUGACAUUUUAUGGCCAUCAAAGUCGCAUAAGACGUCAAAGAAAUAAAAAAAA ..........---......................((((((((.((((........((((((....))))))........(((......))))))).))))))))...... ( -17.20) >DroAna_CAF1 31144 108 - 1 CCCCUUUUUUUUAUGUUUUCCUUUUAUUU-UCAUCUUAUUUUUAUGGCAUCUUAUCCUUUGACAUUUUAUGGCCAUCAAAAUCUCAUAAGACGUCAAUGGGAAGA--AACC .............................-...............((..((((.(((.(((((.((((((((...........)))))))).))))).)))))))--..)) ( -16.90) >consensus CCCCUUUUCU___CGUUUUCCCUUUAUUU_CCAUCUUAUUUUUAUGACGCCUUAUCCCAUGACAUUUUAUGGCCAUCAAAAUCGCAUAAGACGUCAAAGAAAUAA__AAAA ...................................((((((((.(((((.(((((.((((((....)))))).............))))).))))).))))))))...... (-12.04 = -12.27 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:17 2006