
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,470,699 – 11,470,808 |
| Length | 109 |
| Max. P | 0.999227 |

| Location | 11,470,699 – 11,470,808 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.68 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -24.14 |
| Energy contribution | -25.03 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.59 |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.999227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
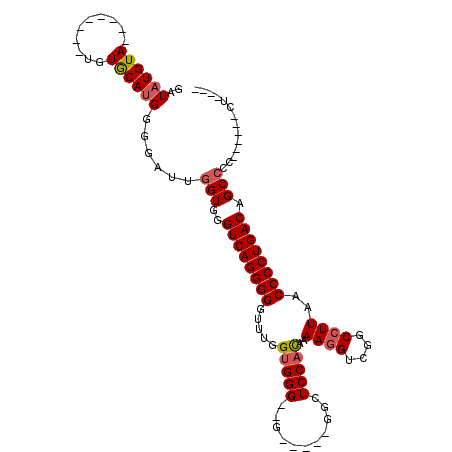
>X_DroMel_CAF1 11470699 109 + 22224390 GAUAUGUAUGUAUGUAUGUACAGGGGAAUUGGUGAGUCAGGGGGGGGGGGGGG--GGUUUUGGCUCCACAAAAAGUCUGCCUUAACCCCUGACAGCACCUCCCCCCUCGUC ....(((((((....)))))))(((((...((((.((((((((..((((.(((--..(((((......)))))..))).))))..))))))))..)))))))))....... ( -49.80) >DroSec_CAF1 15876 85 + 1 GAUAUGUA--------UGUGCAUGGGGAUUGGUGGGUCAGGGGGUUUGGUGGG--G-----GGCUCCAUAAAAGGUCGGCCUUAACCCCUGACAGCCCC------C----- ..(((((.--------...)))))(((...(((..((((((((.....(((((--.-----...)))))..((((....))))..)))))))).)))))------)----- ( -33.90) >DroSim_CAF1 10762 88 + 1 GAUAUGUA--------UGUGCAUGGGGAUUGGUGGGUCAGGGGGUUUAGUGGGGGG-----GGCUCCACAAAAGGUCGGCCUUAACCCCUGACAGCCCC------CU---- ..(((((.--------...)))))(((...(((..((((((((.....((((((..-----..))))))..((((....))))..)))))))).)))))------).---- ( -39.60) >consensus GAUAUGUA________UGUGCAUGGGGAUUGGUGGGUCAGGGGGUUUGGUGGG__G_____GGCUCCACAAAAGGUCGGCCUUAACCCCUGACAGCCCC______CU____ ..((((((..........))))))......(((..((((((((.....(((((...........)))))..((((....))))..)))))))).))).............. (-24.14 = -25.03 + 0.89)


| Location | 11,470,699 – 11,470,808 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.68 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -19.46 |
| Energy contribution | -20.02 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11470699 109 - 22224390 GACGAGGGGGGAGGUGCUGUCAGGGGUUAAGGCAGACUUUUUGUGGAGCCAAAACC--CCCCCCCCCCCCCCUGACUCACCAAUUCCCCUGUACAUACAUACAUACAUAUC .....(((((((((((..((((((((....((......(((((......)))))..--....))....)))))))).))))..)))))))..................... ( -34.80) >DroSec_CAF1 15876 85 - 1 -----G------GGGGCUGUCAGGGGUUAAGGCCGACCUUUUAUGGAGCC-----C--CCCACCAAACCCCCUGACCCACCAAUCCCCAUGCACA--------UACAUAUC -----(------(((..(((((((((..((((....))))...(((....-----.--.)))......)))))))).....)..)))).......--------........ ( -26.30) >DroSim_CAF1 10762 88 - 1 ----AG------GGGGCUGUCAGGGGUUAAGGCCGACCUUUUGUGGAGCC-----CCCCCCACUAAACCCCCUGACCCACCAAUCCCCAUGCACA--------UACAUAUC ----.(------(((..(((((((((..((((....))))..((((....-----....)))).....)))))))).....)..)))).......--------........ ( -29.40) >consensus ____AG______GGGGCUGUCAGGGGUUAAGGCCGACCUUUUGUGGAGCC_____C__CCCACCAAACCCCCUGACCCACCAAUCCCCAUGCACA________UACAUAUC ............((((..((((((((..((((....))))..((((.............)))).....))))))))........))))....................... (-19.46 = -20.02 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:13 2006