
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,461,351 – 11,461,487 |
| Length | 136 |
| Max. P | 0.983880 |

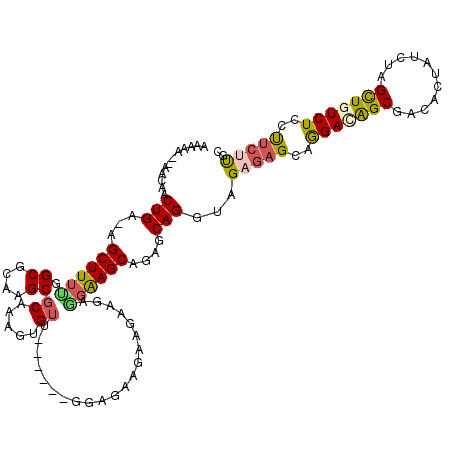
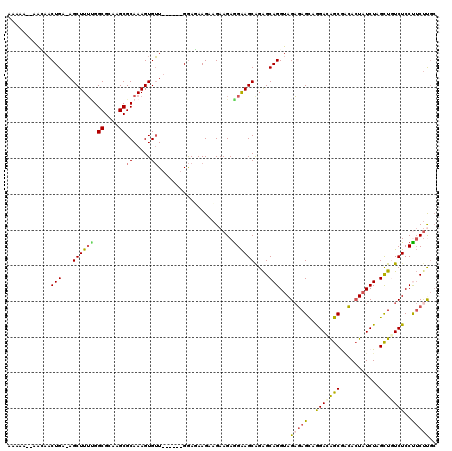
| Location | 11,461,351 – 11,461,463 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -22.08 |
| Energy contribution | -22.44 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11461351 112 + 22224390 AAAAAU-AGGAACUGA-AGCUUUCGGCGCAAGCGCAAAGUGUU------GGAUAAGCAGAAGAGGAAGCAGAGCAGGUAGAGAGCAGGACAGCGGCACUAUCUAGCUGUCUCCCUCUUGC ......-.....(((.-.((((((.((....))......((((------.....)))).....))))))....)))...(((((..(((((((...........)))))))..))))).. ( -33.70) >DroSec_CAF1 6749 111 + 1 AAAAA--AGGAACUGA-AGCUUUUGGCGCAAGCGCAAAGUGUU------GGAUAAGAAGAAGAGGAAGCGGAGCAGGUAGAGAGCAGGACAGCGCCACUAUCUAGCUAUCUCCUUCUUGA .....--.....(..(-.(((((.(((....)).)))))).).------.).(((((((..((((.(((.....((((((.(.((......))..).)))))).))).))))))))))). ( -30.90) >DroEre_CAF1 6534 110 + 1 AAAA---AACAACUGA-AGCUUUUGGCGCAAGCGCAAAGUGUU------GGAGAAGAUGAAGAAGAAGCAGAGCAGGUAGAGAGCAGGACAGCAACACUAUCUAGCUGUCUCCUUCCUGC ....---.....((..-((((((((((....)).))))).)))------..))..............((((.(.(((.(((.(((.(((.((.....)).))).))).)))))).))))) ( -30.90) >DroYak_CAF1 6472 113 + 1 AAAAAGAAACAACUGA-AGCUUUUGGCGCAAGCGCAAAGUGUU------GGAGAAGAAGAAGAGGAAGCAGAGCAGGUAGAGAGCAGGAUAGCGACACUAUCUAGCUGUCUCCUUCUCGC ............((..-((((((((((....)).))))).)))------..))..............((.(((.(((.(((.(((.((((((.....)))))).))).)))))).))))) ( -34.40) >DroAna_CAF1 9971 119 + 1 AUAAAAAAAUAUCUGAAAGCUUGGGGCGCAAGCGCAAAGUGAUAUGGAAGAAGAAGAAGAAGAGGGAGCAGGGCAGGUAGCAAGUGAGAUGGCAAUA-UCUCUAGUUCUCUCCCACUCUC ........(((((.....((((.(.((....)).).)))))))))............(((.(.(((((.(((((.(....)....((((((....))-))))..)))))))))).)))). ( -30.10) >consensus AAAAA__AACAACUGA_AGCUUUUGGCGCAAGCGCAAAGUGUU______GGAGAAGAAGAAGAGGAAGCAGAGCAGGUAGAGAGCAGGACAGCGACACUAUCUAGCUGUCUCCUUCUUGC ............(((...((((((.((....))((.....)).....................))))))....)))...(((((..(((((((...........)))))))..))))).. (-22.08 = -22.44 + 0.36)

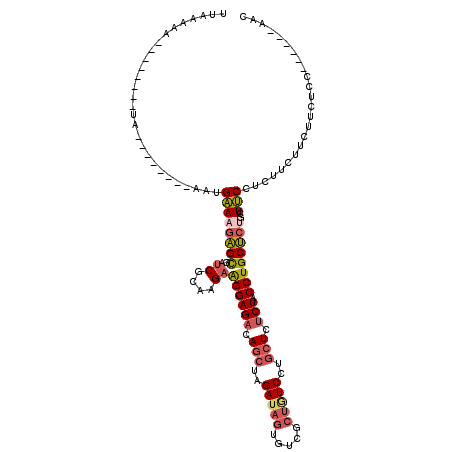
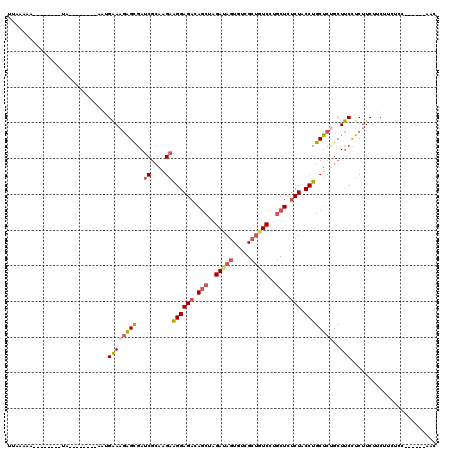
| Location | 11,461,389 – 11,461,487 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.12 |
| Mean single sequence MFE | -26.56 |
| Consensus MFE | -17.86 |
| Energy contribution | -18.62 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11461389 98 - 22224390 UUAAAAA--------UA--------ACUGAAAGAGUGAUCGCAAGAGGGAGACAGCUAGAUAGUGCCGCUGUCCUGCUCUCUACCUGCUCUGCUUCCUCUUCUGCUUAUCC------AAC ......(--------((--------(.(..(((((.((..((((((((.(((.(((..(((((.....)))))..))).))).))).)).))).)))))))..).))))..------... ( -26.70) >DroSec_CAF1 6786 98 - 1 UUAAAAA--------UA--------ACUGAAAGAGUGAUCUCAAGAAGGAGAUAGCUAGAUAGUGGCGCUGUCCUGCUCUCUACCUGCUCCGCUUCCUCUUCUUCUUAUCC------AAC .......--------..--------.....(((((.((.....((.((((((.(((..(((((.....)))))..))).))).))).)).....)))))))..........------... ( -22.60) >DroEre_CAF1 6570 98 - 1 UUAAAAA--------UA--------AAUGAAAGAGCGAUCGCAGGAAGGAGACAGCUAGAUAGUGUUGCUGUCCUGCUCUCUACCUGCUCUGCUUCUUCUUCAUCUUCUCC------AAC .......--------..--------...(((..((((...(((((.(((((.(((...(((((.....))))))))))).)).)))))..))))..)))............------... ( -28.60) >DroYak_CAF1 6511 98 - 1 AUAAAAA--------UA--------AAUGAAAGAGCGAUCGCGAGAAGGAGACAGCUAGAUAGUGUCGCUAUCCUGCUCUCUACCUGCUCUGCUUCCUCUUCUUCUUCUCC------AAC .......--------..--------.....(((((.((..(((((.((((((.(((..(((((.....)))))..))).))).))).))).)).)))))))..........------... ( -27.70) >DroAna_CAF1 10011 119 - 1 CUAAAAACAAAAAAAUAUGUUAAAAAAUGGACUGGCGAUCGAGAGUGGGAGAGAACUAGAGA-UAUUGCCAUCUCACUUGCUACCUGCCCUGCUCCCUCUUCUUCUUCUUCUUCCAUAUC .....((((........)))).....(((((..((.((..(((((.(((((((.....((((-(......)))))....((.....)).)).))))).).))))..)).)).)))))... ( -27.20) >consensus UUAAAAA________UA________AAUGAAAGAGCGAUCGCAAGAAGGAGACAGCUAGAUAGUGUCGCUGUCCUGCUCUCUACCUGCUCUGCUUCCUCUUCUUCUUCUCC______AAC ............................((((((((..((....))((((((.(((..(((((.....)))))..))).))).))))))))..)))........................ (-17.86 = -18.62 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:05 2006