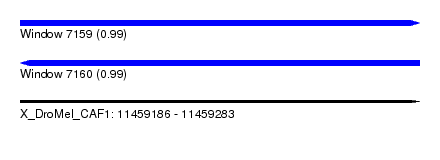
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,459,186 – 11,459,283 |
| Length | 97 |
| Max. P | 0.992668 |

| Location | 11,459,186 – 11,459,283 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.64 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -12.82 |
| Energy contribution | -13.26 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
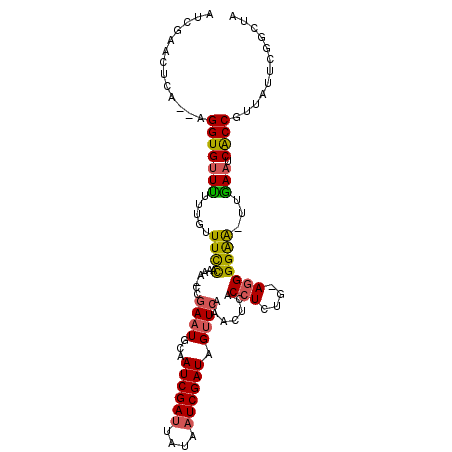
>X_DroMel_CAF1 11459186 97 + 22224390 CAGCCGAAUAACGGCGAAUUAA-CUCCCCU-CAGAGGUGAGUUUGAACUAUCGAUUAUAAUCGAUUGCAUUC-------GGAAACAAAAAACACCU--UGAGUUUGGU ..((((.....))))......(-((...((-((.(((((.(..((....((((((....))))))..))..)-------(....)......)))))--))))...))) ( -26.30) >DroSec_CAF1 4685 106 + 1 UGGCCGAAUAACGGUGAUUCAA-UUCCCCU-CUGAGGUGAGUUUGAACUAUCGAUUAUAAUCGAUAGCAUUCGGAUUUCGGAAACAAAGAACACCUUUUGAGUUCGAU ....((((....((.((.....-.))))((-(.((((((((((((((((((((((....))))))))..))))))))..(....)......))))))..))))))).. ( -30.70) >DroSim_CAF1 4096 104 + 1 UAGCCGAAUAACGGUGAUUCAA-UUCCCCU-CUGAGGUGAGUUUGAACUAUCGAUUAUAAUCGAUAGCAUUCGGAGUUCGGAAACAAAGAACACCU--CGAGUUCGAU ....((((....((.((.....-.))))((-(.((((((..((((((((((((((....))))))))..))))))....(....)......)))))--)))))))).. ( -30.90) >DroEre_CAF1 4627 102 + 1 --GCCAAAUAACGGUGAUUCAAAUCCCCAUCUAGAGGUG-GUUUGAACUAUCGAUUAUAAUCGAUUGUAUUCGG-GUUUGAAAACUUGAAACACCU--UAAAUUGGUU --(((((.....((((..(((((((.((.......)).)-))))))((.((((((....)))))).)).(((((-(((....)))))))).)))).--....))))). ( -28.90) >DroYak_CAF1 4561 93 + 1 CAGCCGUAUAACGGUGGUUCGAAUUCCCCU-UAGAGGUG-GAUUUAACUAUCGAGUAUAAUCGAUUGUAGUCGA-UUUUAAAAAAACCAAACACCU------------ ..((((.....))))((((.....(((((.-....)).)-))(((((..(((((.(((((....))))).))))-).)))))..))))........------------ ( -23.10) >consensus CAGCCGAAUAACGGUGAUUCAA_UUCCCCU_CAGAGGUGAGUUUGAACUAUCGAUUAUAAUCGAUUGCAUUCGG_GUUCGGAAACAAAAAACACCU__UGAGUUCGAU ..((((.....))))...................(((((..........((((((....))))))..............(....)......)))))............ (-12.82 = -13.26 + 0.44)



| Location | 11,459,186 – 11,459,283 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.64 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -14.72 |
| Energy contribution | -14.44 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11459186 97 - 22224390 ACCAAACUCA--AGGUGUUUUUUGUUUCC-------GAAUGCAAUCGAUUAUAAUCGAUAGUUCAAACUCACCUCUG-AGGGGAG-UUAAUUCGCCGUUAUUCGGCUG .((...((((--(((((......((((..-------((((...((((((....)))))).)))))))).))))).))-)).))..-.......((((.....)))).. ( -27.50) >DroSec_CAF1 4685 106 - 1 AUCGAACUCAAAAGGUGUUCUUUGUUUCCGAAAUCCGAAUGCUAUCGAUUAUAAUCGAUAGUUCAAACUCACCUCAG-AGGGGAA-UUGAAUCACCGUUAUUCGGCCA .(((((.......(((((((...((((((.......((..(((((((((....)))))))))))...(((......)-)))))))-).))).))))....)))))... ( -29.60) >DroSim_CAF1 4096 104 - 1 AUCGAACUCG--AGGUGUUCUUUGUUUCCGAACUCCGAAUGCUAUCGAUUAUAAUCGAUAGUUCAAACUCACCUCAG-AGGGGAA-UUGAAUCACCGUUAUUCGGCUA .(((((....--.(((((((...((((((.......((..(((((((((....)))))))))))...(((......)-)))))))-).))).))))....)))))... ( -30.80) >DroEre_CAF1 4627 102 - 1 AACCAAUUUA--AGGUGUUUCAAGUUUUCAAAC-CCGAAUACAAUCGAUUAUAAUCGAUAGUUCAAAC-CACCUCUAGAUGGGGAUUUGAAUCACCGUUAUUUGGC-- ..((((....--.((((.((((((((..((...-..((((...((((((....)))))).))))....-...(....).))..)))))))).)))).....)))).-- ( -27.80) >DroYak_CAF1 4561 93 - 1 ------------AGGUGUUUGGUUUUUUUAAAA-UCGACUACAAUCGAUUAUACUCGAUAGUUAAAUC-CACCUCUA-AGGGGAAUUCGAACCACCGUUAUACGGCUG ------------.(((((((((...........-..(((((...((((......)))))))))...((-(.((....-.)))))..))))).))))............ ( -19.20) >consensus AUCGAACUCA__AGGUGUUUUUUGUUUCCAAAA_CCGAAUGCAAUCGAUUAUAAUCGAUAGUUCAAACUCACCUCUG_AGGGGAA_UUGAAUCACCGUUAUUCGGCUA .............(((((((.....((((.......((((...((((((....)))))).)))).......(((....)))))))...))).))))............ (-14.72 = -14.44 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:02 2006