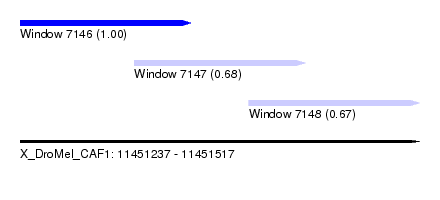
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,451,237 – 11,451,517 |
| Length | 280 |
| Max. P | 0.995222 |

| Location | 11,451,237 – 11,451,357 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -41.65 |
| Consensus MFE | -40.22 |
| Energy contribution | -39.98 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11451237 120 + 22224390 UGCGAACAGGAAGUCGUAGCUACCGAUAAAACGGCCAUCAAUACAAUCUUGGAAGCUGAUGUGCGUCGCACAGAGAUGCUAAAGAAGGCCGAUGAACUGGAGAAACAGUUUGUCGCUGGC (((((........)))))((((.((((....(((((.((...(((.((..(....).)))))(((((.......)))))....)).)))))..((((((......)))))))))).)))) ( -36.10) >DroSec_CAF1 5168 120 + 1 UGCGAACAGGAAGUCGUAGCUACCGAUAAAACGGCCAUCAAUACAAUCCUGGAAGCGGAUGUGCGUCGCACAGAGAUGCUAAAGAAGGCCGAUGAACUGGAGAAACAGUUUGUCGGUGGC (((((........)))))(((((((((....(((((.((...(((.(((.(....)))))))(((((.......)))))....)).)))))..((((((......))))))))))))))) ( -45.80) >DroEre_CAF1 8527 120 + 1 UGCGAACAGGAAGUCGUAGCUACCGACAAAACGGCCAUCGAUACAAUCCUGGAAGCGGAUGUGCGACGCACACAGAUGCUAAAGAAGGCCGAUGAGUUGGAGAAACAGUUUGUCGGUGGC (((((........)))))((((((((((((.(((((.(((.((((.(((.(....))))))))))).(((......))).......)))))....(((.....)))..)))))))))))) ( -44.10) >DroYak_CAF1 8699 120 + 1 UGCGAACAGGAAGUCGUUGCUACCGACAAAACGGCCAUCGAUACAAUCCUGGAAGCGGAUGUGCGACGCACACAGAUGCUAAAGAAGGCUGAUGAACUGGAGAAACAGUUUGUUGGUGGC .((((........)))).(((((((((....(((((.(((.((((.(((.(....))))))))))).(((......))).......)))))..((((((......))))))))))))))) ( -40.60) >consensus UGCGAACAGGAAGUCGUAGCUACCGACAAAACGGCCAUCAAUACAAUCCUGGAAGCGGAUGUGCGACGCACACAGAUGCUAAAGAAGGCCGAUGAACUGGAGAAACAGUUUGUCGGUGGC (((((........)))))(((((((((....(((((.((....((....))..((((..(((((...)))))....))))...)).)))))..((((((......))))))))))))))) (-40.22 = -39.98 + -0.25)

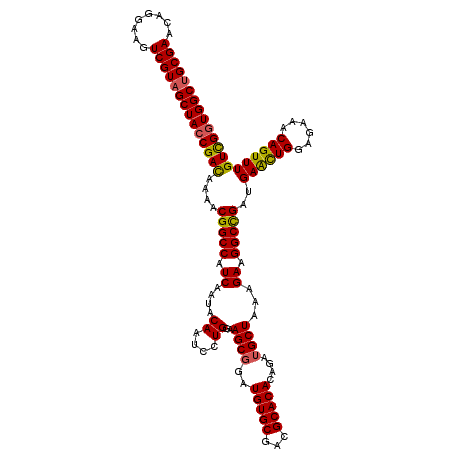
| Location | 11,451,317 – 11,451,437 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.14 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -35.80 |
| Energy contribution | -35.67 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.680727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11451317 120 + 22224390 AAAGAAGGCCGAUGAACUGGAGAAACAGUUUGUCGCUGGCGAUUUGACGGUGCAGGAGGAGCUAAACGAUACCUUCGCGGAACUGAAAGCCAUUGGAGCCUACUCGGCGGAAGCCAGGGC .....(((((((..(((((......)))))..))).((((.......((((.(.(((((..(.....)...)))))...).))))...)))).....)))).((((((....))).))). ( -39.90) >DroSec_CAF1 5248 120 + 1 AAAGAAGGCCGAUGAACUGGAGAAACAGUUUGUCGGUGGCGAUUUGACUGUGCAGGAGGAGCUAAACGAUACCUUCGCGGAACUGAAAGCCAUUGGAGCCUACUCGGCGGAAGCCAGGGC .....(((((((..(((((......)))))..)))(((((..((((..(.(((.(((((..(.....)...)))))))).)..)))).)))))....)))).((((((....))).))). ( -42.70) >DroEre_CAF1 8607 120 + 1 AAAGAAGGCCGAUGAGUUGGAGAAACAGUUUGUCGGUGGCGAUUUAACUGUGCAGGAGGAGCUAAACGAUACCUUCGCAGAACUGAAAGCAAUUGGAGCGUACUCGGCGGAAGCCAGAGC .......(((((..(..((......))..)..))))).(((.(((((.(((.(((..((((.((.....)).))))......)))...))).))))).))).((((((....))).))). ( -34.90) >DroYak_CAF1 8779 120 + 1 AAAGAAGGCUGAUGAACUGGAGAAACAGUUUGUUGGUGGCGAUUUGACUGUGCAGGAGGAGCUAAACGAUACCUUCGCGGAACUGAAAGCCAUUGGAGCGUACUCGGCGGAAGCCAGAGC .......(((...((((((......)))))).(..(((((..((((..(.(((.(((((..(.....)...)))))))).)..)))).)))))..))))...((((((....))).))). ( -37.30) >consensus AAAGAAGGCCGAUGAACUGGAGAAACAGUUUGUCGGUGGCGAUUUGACUGUGCAGGAGGAGCUAAACGAUACCUUCGCGGAACUGAAAGCCAUUGGAGCCUACUCGGCGGAAGCCAGAGC .....(((((((..(((((......)))))..)))(((((..((((..(.(((.(((((..(.....)...)))))))).)..)))).)))))....)))).((((((....))).))). (-35.80 = -35.67 + -0.12)



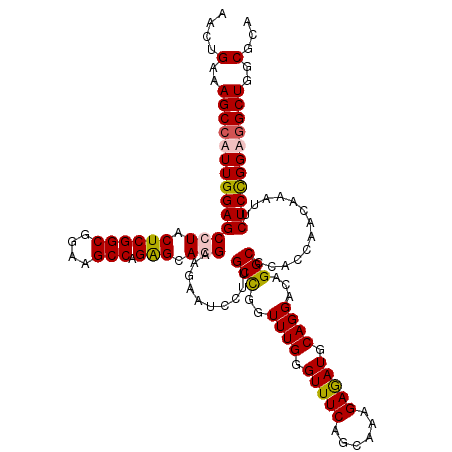
| Location | 11,451,397 – 11,451,517 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.19 |
| Mean single sequence MFE | -41.65 |
| Consensus MFE | -38.45 |
| Energy contribution | -38.83 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11451397 120 + 22224390 AACUGAAAGCCAUUGGAGCCUACUCGGCGGAAGCCAGGGCACGAAGAAUCCUUGCCGGUUUGGGUUUCAGCAAAGAGAUGCAGGACAGGCCGACGAACAAAUUCUCCGGAGGCUGGCGCA ........(((..(((((((((..((((....)))..((((.(.......).)))).)..)))))))))(((......)))......((((..((...........))..)))))))... ( -40.40) >DroSec_CAF1 5328 120 + 1 AACUGAAAGCCAUUGGAGCCUACUCGGCGGAAGCCAGGGCACGCAGAAUCCUUGCCGGUUUGGGUUUCAGCAAAGAGAUGCAGGACAGGCCGACGAACAAAUUCUCCGGAGGCUGGCGCA ........(((..(((((((((..((((....)))..((((...((....)))))).)..)))))))))(((......)))......((((..((...........))..)))))))... ( -39.70) >DroEre_CAF1 8687 120 + 1 AACUGAAAGCAAUUGGAGCGUACUCGGCGGAAGCCAGAGCACGAAGAAUCCUUGCCGGUUUGGGUUUCAGCAAAGAAAUGCAGGAUAGGCCCACCAACAAAUUCUCUGGUGGCUGGCGCA ........((....(((.(((.((((((....))).))).))).....)))..(((((((((.(((((......))))).))))......((((((..........))))))))))))). ( -42.20) >DroYak_CAF1 8859 120 + 1 AACUGAAAGCCAUUGGAGCGUACUCGGCGGAAGCCAGAGCACGACGAAUUCUUGCUGGUUUGGGUUUCAGCAAAGAGAUGCAGGAUAGGCCCACCAAUAAAUUCUCUGGUGGCUGGCGCA ....(..((((((..((((((.((((((....))).))).)))........(((.((((..(((((((.(((......)))..))..))))))))).)))...)))..))))))..)... ( -44.30) >consensus AACUGAAAGCCAUUGGAGCCUACUCGGCGGAAGCCAGAGCACGAAGAAUCCUUGCCGGUUUGGGUUUCAGCAAAGAGAUGCAGGACAGGCCCACCAACAAAUUCUCCGGAGGCUGGCGCA ....(..((((((((((((((.((((((....))).))).)))..........(((..((((.(((((......))))).))))...))).............)))))))))))..)... (-38.45 = -38.83 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:50 2006