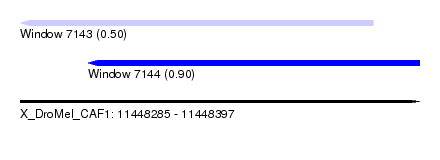
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,448,285 – 11,448,397 |
| Length | 112 |
| Max. P | 0.904690 |

| Location | 11,448,285 – 11,448,384 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.73 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -16.33 |
| Energy contribution | -17.77 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.50 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11448285 99 - 22224390 AACAAGGUUCCAUUGGUGAACAGAGUGCCGGCGUGGGACAAAACAUUAUUGAGGGCGACUUGCUGGUGUUAAAUGCCAAUUCCACAAACGUGGAUCUU---------------------U ...((((....((((((......((..((((((((........))).....(((....))))))))..))....))))))(((((....)))))))))---------------------. ( -25.60) >DroSec_CAF1 1968 99 - 1 GAAAAGGUUCCAUCGGUAAACAGAGUGCCGGCAUAGGACGCAACAUUAUUGACGGCGACCUGCCGGUGUCCAAUGCCAAUGCCACAAACGCGAAUCUU---------------------U ..(((((((((...((((.......))))(((((.((((((..((....)).((((.....)))))))))).)))))............).)))))))---------------------) ( -35.30) >DroSim_CAF1 1911 93 - 1 GAAAAGGUUCCAUCGGUAAACAGAGUGCCGGCAUAGGACGCAACAUUAUUUACGGCGACCUGCCGGUGUCCAAUGACACUGCA------UUGAAUCUU---------------------U ..((((((((.((((((.........(((((((..((.(((.............))).)))))))))(((....))))))).)------).)))))))---------------------) ( -28.62) >DroYak_CAF1 5371 120 - 1 GAAAGGGUUCCGUUGGUGGAGAGAGUGCCAGAGUAGGACCAAGCAAUGUACGCGACCGCCUGCUGGUGCACAAUGACAAUUCCGCGUUCGGGAAUCUUAGUGUCAUCGAUGGCUCUCUUU .((((((..((((((((((.(((((((((..((((((.....((.......)).....)))))).).))))........(((((....))))).))))....))))))))))..)))))) ( -41.80) >consensus GAAAAGGUUCCAUCGGUAAACAGAGUGCCGGCAUAGGACGAAACAUUAUUGACGGCGACCUGCCGGUGUCCAAUGACAAUGCCACAAACGUGAAUCUU_____________________U ...(((((((((((((((......(..(((((..(((.(((.............))).))))))))..)....))))))))..........)))))))...................... (-16.33 = -17.77 + 1.44)


| Location | 11,448,304 – 11,448,397 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.80 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.52 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11448304 93 - 22224390 ---------------------------GGUGCUAUAGAUGAACAAGGUUCCAUUGGUGAACAGAGUGCCGGCGUGGGACAAAACAUUAUUGAGGGCGACUUGCUGGUGUUAAAUGCCAAU ---------------------------((..((......((((...))))..(((.....)))))..))(((((..((((...((....))(((....))).....))))..)))))... ( -21.80) >DroSec_CAF1 1987 113 - 1 -------UAGAUGGAAAAGGUGCUUUAGGUGCUAUAGAUGGAAAAGGUUCCAUCGGUAAACAGAGUGCCGGCAUAGGACGCAACAUUAUUGACGGCGACCUGCCGGUGUCCAAUGCCAAU -------...........((..((((...((((...(((((((....)))))))))))...))))..))(((((.((((((..((....)).((((.....)))))))))).)))))... ( -45.90) >DroSim_CAF1 1924 120 - 1 GGUGCUAUAGAUGGAAAAGGUGCUAUAGGUGCUAUAGAUGGAAAAGGUUCCAUCGGUAAACAGAGUGCCGGCAUAGGACGCAACAUUAUUUACGGCGACCUGCCGGUGUCCAAUGACACU (((((..(((((((.....(((((....(((((...(((((((....)))))))((((.......))))))))).)).)))....)))))))..)).)))....((((((....)))))) ( -37.50) >DroYak_CAF1 5411 100 - 1 --------------------UGGUCGUCGUGCUACAGAUGGAAAGGGUUCCGUUGGUGGAGAGAGUGCCAGAGUAGGACCAAGCAAUGUACGCGACCGCCUGCUGGUGCACAAUGACAAU --------------------..(((((.((.((((.(((((((....))))))).)))).....(..(((..(((((.....((.......)).....))))))))..))).)))))... ( -33.50) >consensus ____________________UGCU_UAGGUGCUAUAGAUGGAAAAGGUUCCAUCGGUAAACAGAGUGCCGGCAUAGGACGAAACAUUAUUGACGGCGACCUGCCGGUGUCCAAUGACAAU ................................(((.(((((((....))))))).)))......(..(((((..(((.(((.............))).))))))))..)........... (-18.40 = -18.52 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:46 2006