
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,424,994 – 11,425,125 |
| Length | 131 |
| Max. P | 0.988963 |

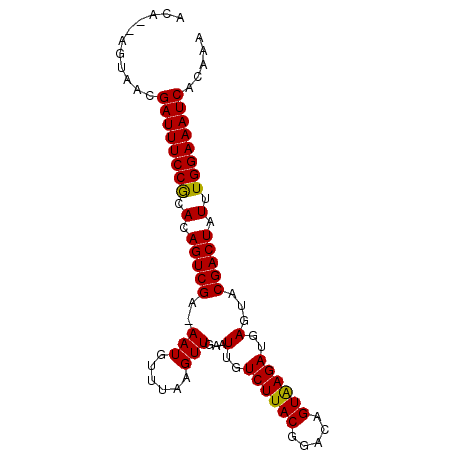
| Location | 11,424,994 – 11,425,085 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -24.41 |
| Energy contribution | -23.97 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.21 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11424994 91 + 22224390 GCAAGAGUAACGAUUUCCGCAUAGUCGAAAAUGUUUAAGUUGAAUUGUCUUACGGACAGUGAGAUGAGUACGACUAUUUGGAAAUCACAAA ...........((((((((.(((((((..(((......)))...(..((((((.....))))))..)...))))))).))))))))..... ( -28.10) >DroEre_CAF1 5682 88 + 1 ACA--AGUAACGAUUUCCACACAGUCGA-AAUGUUUAAGUUGAAUUGUCUUACGGACAGUAAGAUGAGUACGACUAUUUGGAAAUCACAAA ...--......((((((((.(.(((((.-(((......)))...(..((((((.....))))))..)...))))).).))))))))..... ( -24.60) >DroYak_CAF1 5694 88 + 1 ACA--AGUAACGAUUUCCGAACAGUCGA-AAUGUUGAAGUUGAAUUGUCUUACGGACAGUAAGAUGAGUACGACUAUUUGGAAAUCACAAA ...--......((((((((((.(((((.-(((......)))...(..((((((.....))))))..)...))))).))))))))))..... ( -27.80) >consensus ACA__AGUAACGAUUUCCGCACAGUCGA_AAUGUUUAAGUUGAAUUGUCUUACGGACAGUAAGAUGAGUACGACUAUUUGGAAAUCACAAA ...........((((((((.(.(((((..(((......)))...(..((((((.....))))))..)...))))).).))))))))..... (-24.41 = -23.97 + -0.44)


| Location | 11,424,994 – 11,425,085 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -21.11 |
| Consensus MFE | -19.76 |
| Energy contribution | -19.87 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11424994 91 - 22224390 UUUGUGAUUUCCAAAUAGUCGUACUCAUCUCACUGUCCGUAAGACAAUUCAACUUAAACAUUUUCGACUAUGCGGAAAUCGUUACUCUUGC .....(((((((..(((((((............((((.....))))..................)))))))..)))))))........... ( -18.31) >DroEre_CAF1 5682 88 - 1 UUUGUGAUUUCCAAAUAGUCGUACUCAUCUUACUGUCCGUAAGACAAUUCAACUUAAACAUU-UCGACUGUGUGGAAAUCGUUACU--UGU .....((((((((.(((((((......((((((.....))))))..................-.))))))).))))))))......--... ( -22.81) >DroYak_CAF1 5694 88 - 1 UUUGUGAUUUCCAAAUAGUCGUACUCAUCUUACUGUCCGUAAGACAAUUCAACUUCAACAUU-UCGACUGUUCGGAAAUCGUUACU--UGU .....(((((((.((((((((......((((((.....))))))..................-.)))))))).)))))))......--... ( -22.21) >consensus UUUGUGAUUUCCAAAUAGUCGUACUCAUCUUACUGUCCGUAAGACAAUUCAACUUAAACAUU_UCGACUGUGCGGAAAUCGUUACU__UGU .....(((((((..(((((((......((((((.....))))))....................)))))))..)))))))........... (-19.76 = -19.87 + 0.11)



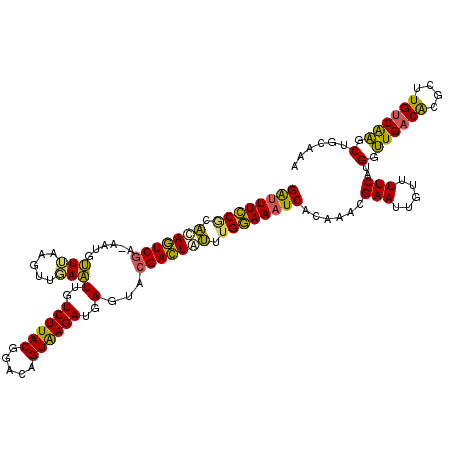
| Location | 11,425,005 – 11,425,125 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.20 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -27.51 |
| Energy contribution | -26.08 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11425005 120 + 22224390 GAUUUCCGCAUAGUCGAAAAUGUUUAAGUUGAAUUGUCUUACGGACAGUGAGAUGAGUACGACUAUUUGGAAAUCACAAACGAAUUGUUUUCAUGGUUGACGCGCUUGUCAAGCUACAAA ((((((((.(((((((..(((......)))...(..((((((.....))))))..)...))))))).))))))))......(((.....)))..(.((((((....)))))).)...... ( -34.40) >DroEre_CAF1 5691 119 + 1 GAUUUCCACACAGUCGA-AAUGUUUAAGUUGAAUUGUCUUACGGACAGUAAGAUGAGUACGACUAUUUGGAAAUCACAAACGAAUUGUUUUCAUGGUUGACACGCUUGCCAAGCUGCAAA ((((((((.(.(((((.-(((......)))...(..((((((.....))))))..)...))))).).)))))))).........((((...(.((((..........)))).)..)))). ( -29.20) >DroYak_CAF1 5703 119 + 1 GAUUUCCGAACAGUCGA-AAUGUUGAAGUUGAAUUGUCUUACGGACAGUAAGAUGAGUACGACUAUUUGGAAAUCACAAACGAAUUGUUUUCAUGAUUGACACGCUUGUCAAGCUGCAAA ((((((((((.(((((.-(((......)))...(..((((((.....))))))..)...))))).))))))))))......(((.....))).((.((((((....))))))....)).. ( -33.50) >DroAna_CAF1 6535 116 + 1 GGCUUUCUCUCAAUCAA-AAUGAUAAAUUUUAUUCGUCUUACGGAUAGUGAGAAAAAUAAGAUUGAUUGAAAAUCAU---UGAAAUGUUUUCUGGUAUGACACGGAUGUCGUGCUGUGAA (..((((..((((((((-(((.....)))))(((..((((((.....))))))..)))..))))))..))))..)..---.........(((((((((((((....)))))))))).))) ( -29.60) >consensus GAUUUCCGCACAGUCGA_AAUGUUUAAGUUGAAUUGUCUUACGGACAGUAAGAUGAGUACGACUAUUUGGAAAUCACAAACGAAUUGUUUUCAUGGUUGACACGCUUGUCAAGCUGCAAA ((((((((.(((((((......(((.....)))(..((((((.....))))))..)...))))))).))))))))......(((.....)))..(.((((((....)))))).)...... (-27.51 = -26.08 + -1.44)


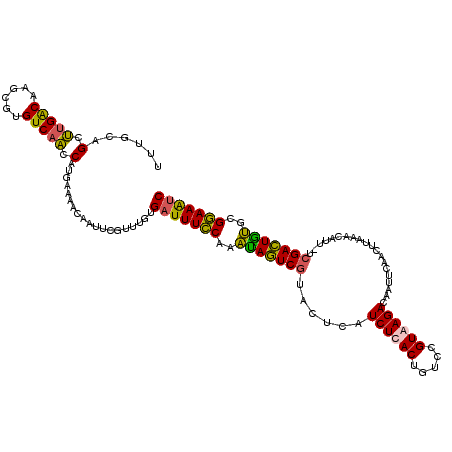
| Location | 11,425,005 – 11,425,125 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.20 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -20.22 |
| Energy contribution | -19.22 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11425005 120 - 22224390 UUUGUAGCUUGACAAGCGCGUCAACCAUGAAAACAAUUCGUUUGUGAUUUCCAAAUAGUCGUACUCAUCUCACUGUCCGUAAGACAAUUCAACUUAAACAUUUUCGACUAUGCGGAAAUC ((..(.(.(((((......)))))).)..)).((((.....))))(((((((..(((((((............((((.....))))..................)))))))..))))))) ( -22.51) >DroEre_CAF1 5691 119 - 1 UUUGCAGCUUGGCAAGCGUGUCAACCAUGAAAACAAUUCGUUUGUGAUUUCCAAAUAGUCGUACUCAUCUUACUGUCCGUAAGACAAUUCAACUUAAACAUU-UCGACUGUGUGGAAAUC ...........(((((((..(((....)))........)))))))((((((((.(((((((......((((((.....))))))..................-.))))))).)))))))) ( -27.51) >DroYak_CAF1 5703 119 - 1 UUUGCAGCUUGACAAGCGUGUCAAUCAUGAAAACAAUUCGUUUGUGAUUUCCAAAUAGUCGUACUCAUCUUACUGUCCGUAAGACAAUUCAACUUCAACAUU-UCGACUGUUCGGAAAUC ...((((.((((((....)))))).)(((((.....))))).)))(((((((.((((((((......((((((.....))))))..................-.)))))))).))))))) ( -27.31) >DroAna_CAF1 6535 116 - 1 UUCACAGCACGACAUCCGUGUCAUACCAGAAAACAUUUCA---AUGAUUUUCAAUCAAUCUUAUUUUUCUCACUAUCCGUAAGACGAAUAAAAUUUAUCAUU-UUGAUUGAGAGAAAGCC .(((..(((((.....))))).......(((.....))).---.)))(((((((((((..((((((.(((.((.....)).))).))))))...........-)))))))))))...... ( -18.52) >consensus UUUGCAGCUUGACAAGCGUGUCAACCAUGAAAACAAUUCGUUUGUGAUUUCCAAAUAGUCGUACUCAUCUCACUGUCCGUAAGACAAUUCAACUUAAACAUU_UCGACUGUGCGGAAAUC ......(.(((((......))))).)...................(((((((..(((((((......((((((.....))))))....................)))))))..))))))) (-20.22 = -19.22 + -1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:40 2006