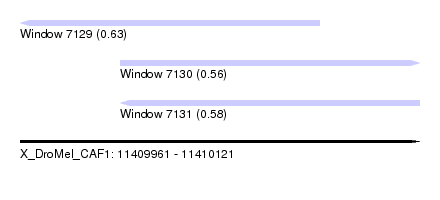
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,409,961 – 11,410,121 |
| Length | 160 |
| Max. P | 0.634084 |

| Location | 11,409,961 – 11,410,081 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -45.47 |
| Consensus MFE | -33.73 |
| Energy contribution | -33.77 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11409961 120 - 22224390 ACGUUGGGCAACAGUAUGCUAGGUGGCGCCGCUAUGUUCAUUGGCGGUGGAUCGACACCGAGCAUGACGCCACCGAUGACGCCGUGGGCUAACUGCAACACGCCGCGAUACUUCUCUCCA .((((((.......((((((.((((.((((((((.......))))))))......)))).))))))......)))))).(((.(((.((.....))....))).)))............. ( -42.12) >DroVir_CAF1 1188 120 - 1 UCCUUGGGCAGCAGCAUGCUGGGCGGCGCUGCCAUCUUUAUGGGCGGCGGCUCCACGCCCAGCAUGACGCCGCCGAUGACGCCUUGGGUGAACUGCCAUACGCCGCGCUACUUCUCACCC ......(((.((..(((((((((((..((((((..((....))..))))))....)))))))))))..)).)))...........((((((...(((.......).))......)))))) ( -53.70) >DroGri_CAF1 1221 120 - 1 UCCCUUGGUAGCAGUAUGCUUGGCGGUGCUGCCAUCUUUAUGGGCGGCGGUUCCACGCCCAGCAUGACGCCACCAAUGACGCCGUGGGUCAAUUGCCAUACGCCGCGCUACUUCUCACCG ......(((((.((((.((.(((((((((((((((....))))..((((......))))))))))(((.((((..........)))))))..........))))).)))))).)).))). ( -44.80) >DroWil_CAF1 1196 120 - 1 ACACUGGGCAGCGCAAUGAUGGGCGGUGCUGCCAUGUUUAUUGGAGGCGGUUCCACGCCUAGCAUGACACCGCCAAUGACACCUUGGGUGAACUGUAAUACACCGAGGUACUUCUCUCCG ......((((((((...........)))))))).........(((((((((..((.((...)).))..))))))......((((((((((........))).))))))).......))). ( -42.30) >DroMoj_CAF1 1245 120 - 1 UCUCUUGGCAGCAGCAUGCUGGGCGGUGCUGCCAUCUUUAUGGGCGGUGGCUCCACGCCAAGUAUGACGCCGCCAAUGACGCCAUGGGUGAAUUGCCAUACACCCCGUUACUUUUCGCCA ....(((((.((..((((((.((((..((..((..((....))..))..))....)))).))))))..)).)))))(((((....(((((..........)))))))))).......... ( -46.10) >DroPer_CAF1 6744 120 - 1 ACCCUGGGAAGCAGCAUGCUGGGCGGCACGGCCAUGUUCAUUGGCGGUGGCUCUACGCCGAGCAUGACGCCUCCGAUGACACCGUGGGUGAACUGCAAUACGCCACGCUACUNNNNNNNN ......(((.((..((((((.((((.(((.((((.......)))).)))......)))).))))))..)).)))........((((((((........))).)))))............. ( -43.80) >consensus ACCCUGGGCAGCAGCAUGCUGGGCGGCGCUGCCAUCUUUAUGGGCGGCGGCUCCACGCCCAGCAUGACGCCGCCAAUGACGCCGUGGGUGAACUGCAAUACGCCGCGCUACUUCUCACCA ......(((.((..((((((.((((.(((((((.........)))))))......)))).))))))..)).)))........((((((((........))).)))))............. (-33.73 = -33.77 + 0.03)


| Location | 11,410,001 – 11,410,121 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -49.32 |
| Consensus MFE | -32.17 |
| Energy contribution | -32.78 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11410001 120 + 22224390 GUCAUCGGUGGCGUCAUGCUCGGUGUCGAUCCACCGCCAAUGAACAUAGCGGCGCCACCUAGCAUACUGUUGCCCAACGUAUUGGGAAUCUCGAUGCCGAAACGACACUUCUCUGCAUCG (((...(((((((((..((((((((......)))))...........))))))))))))..((((...(...(((((....)))))...)...))))......))).............. ( -39.30) >DroVir_CAF1 1228 120 + 1 GUCAUCGGCGGCGUCAUGCUGGGCGUGGAGCCGCCGCCCAUAAAGAUGGCAGCGCCGCCCAGCAUGCUGCUGCCCAAGGAGUUGGGUAUCUCGAUGCCGAAACGGCAUUUCUCGGCGUCC ......(((((((.(((((((((((.(..((.((((.(......).)))).)).)))))))))))).)))))))...(((((((((......(((((((...))))))).)))))).))) ( -60.00) >DroGri_CAF1 1261 120 + 1 GUCAUUGGUGGCGUCAUGCUGGGCGUGGAACCGCCGCCCAUAAAGAUGGCAGCACCGCCAAGCAUACUGCUACCAAGGGAAUUGGGUAUCUCGAUGCCGAAACGGCACUUCUCGGCGUCC (((.(((((((((..((((((((((((....)).))))).......((((......)))))))))..)))))))))((((((((((...)))))(((((...))))).)))))))).... ( -50.40) >DroWil_CAF1 1236 120 + 1 GUCAUUGGCGGUGUCAUGCUAGGCGUGGAACCGCCUCCAAUAAACAUGGCAGCACCGCCCAUCAUUGCGCUGCCCAGUGUGUUCGGUAUCUCAAUGCCGAAUCGACAUUUCUCGGCAUCG (((...((((((..(((((...)))))..))))))........((((((((((...((........))))))))..))))(((((((((....))))))))).))).............. ( -43.60) >DroMoj_CAF1 1285 120 + 1 GUCAUUGGCGGCGUCAUACUUGGCGUGGAGCCACCGCCCAUAAAGAUGGCAGCACCGCCCAGCAUGCUGCUGCCAAGAGAAUUGGGAAUCUCAAUGCCAAAGCGACACUUCUCCGCGUCC ......((((((......((((((((((.((....))))))......(((((((..((...)).)))))))))))))...((((((...)))))))))...(((.........)))))). ( -40.10) >DroAna_CAF1 8672 120 + 1 GUCAUCGGAGGCGUCAUGCUCGGCGUGGAGCCGCCGCCAAUGAACAUGGCGGCGCCGCCCAGCAUGCUACUGCCCAGCGUGUUGGGGAUCUCGAUGCCGAAGCGGCACUUCUCGGCGUCG (.(..((((((.(((..(((((((((.(((.((((((((.......))))))))...(((((((((((.......)))))))))))...))).)))))).)))))).)))).))..).). ( -62.50) >consensus GUCAUCGGCGGCGUCAUGCUCGGCGUGGAGCCGCCGCCAAUAAACAUGGCAGCACCGCCCAGCAUGCUGCUGCCCAGCGAAUUGGGUAUCUCGAUGCCGAAACGACACUUCUCGGCGUCC ......(((((((..(((((.((((((....))).(((.........)))......))).)))))..)))))))..................((((((((...........)))))))). (-32.17 = -32.78 + 0.62)


| Location | 11,410,001 – 11,410,121 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -49.17 |
| Consensus MFE | -36.82 |
| Energy contribution | -35.85 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11410001 120 - 22224390 CGAUGCAGAGAAGUGUCGUUUCGGCAUCGAGAUUCCCAAUACGUUGGGCAACAGUAUGCUAGGUGGCGCCGCUAUGUUCAUUGGCGGUGGAUCGACACCGAGCAUGACGCCACCGAUGAC ((((((.((((.......)))).))))))............((((((.......((((((.((((.((((((((.......))))))))......)))).))))))......)))))).. ( -43.32) >DroVir_CAF1 1228 120 - 1 GGACGCCGAGAAAUGCCGUUUCGGCAUCGAGAUACCCAACUCCUUGGGCAGCAGCAUGCUGGGCGGCGCUGCCAUCUUUAUGGGCGGCGGCUCCACGCCCAGCAUGACGCCGCCGAUGAC ....(((((((.......)))))))((((((........)))....(((.((..(((((((((((..((((((..((....))..))))))....)))))))))))..)).))))))... ( -58.10) >DroGri_CAF1 1261 120 - 1 GGACGCCGAGAAGUGCCGUUUCGGCAUCGAGAUACCCAAUUCCCUUGGUAGCAGUAUGCUUGGCGGUGCUGCCAUCUUUAUGGGCGGCGGUUCCACGCCCAGCAUGACGCCACCAAUGAC .((.(((((((.......))))))).))..............(.(((((.((..((((((.((((..((((((..((....))..))))))....)))).))))))..)).))))).).. ( -47.20) >DroWil_CAF1 1236 120 - 1 CGAUGCCGAGAAAUGUCGAUUCGGCAUUGAGAUACCGAACACACUGGGCAGCGCAAUGAUGGGCGGUGCUGCCAUGUUUAUUGGAGGCGGUUCCACGCCUAGCAUGACACCGCCAAUGAC ((((((((((.........)))))))))).....((((..(((...((((((((...........)))))))).)))...)))).((((((..((.((...)).))..))))))...... ( -44.80) >DroMoj_CAF1 1285 120 - 1 GGACGCGGAGAAGUGUCGCUUUGGCAUUGAGAUUCCCAAUUCUCUUGGCAGCAGCAUGCUGGGCGGUGCUGCCAUCUUUAUGGGCGGUGGCUCCACGCCAAGUAUGACGCCGCCAAUGAC .(((((......)))))((....))...((((........))))(((((.((..((((((.((((..((..((..((....))..))..))....)))).))))))..)).))))).... ( -49.20) >DroAna_CAF1 8672 120 - 1 CGACGCCGAGAAGUGCCGCUUCGGCAUCGAGAUCCCCAACACGCUGGGCAGUAGCAUGCUGGGCGGCGCCGCCAUGUUCAUUGGCGGCGGCUCCACGCCGAGCAUGACGCCUCCGAUGAC ....(((((..((.....)))))))((((((...((((......))))..((..((((((.((((.((((((((.......))))))))......)))).))))))..)))).))))... ( -52.40) >consensus CGACGCCGAGAAGUGCCGUUUCGGCAUCGAGAUACCCAACACCCUGGGCAGCAGCAUGCUGGGCGGCGCUGCCAUCUUUAUGGGCGGCGGCUCCACGCCCAGCAUGACGCCGCCAAUGAC .((.((((((.........)))))).))..................(((.((..((((((.((((..((((((.((.....))..))))))....)))).))))))..)).)))...... (-36.82 = -35.85 + -0.97)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:34 2006