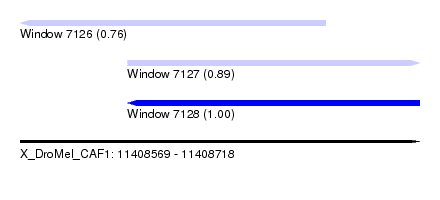
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,408,569 – 11,408,718 |
| Length | 149 |
| Max. P | 0.998467 |

| Location | 11,408,569 – 11,408,683 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.08 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -21.18 |
| Energy contribution | -21.26 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11408569 114 - 22224390 -GGCGGUCAAGCCGUAGUUAAGUACCGACUACGCUCGAUCGCCC----GA-ACGGGAAAGGUGAAUUUAAUAAUAUUAUGUUUGUAUACAGCAAAUAGGUCUCUCAUACAAAAUUCAAUA -(((((((.(((.((((((.......))))))))).))))))).----..-..........(((((((..........(((((((.....))))))).............)))))))... ( -29.00) >DroSec_CAF1 1658 115 - 1 -GGCGGUCGAGCCGUAGUUAAGUACCGACUACGCUUGACCGCCC----GAAACGGGAAAGGUGAAUAUAAUAACAUCUAGUUUUUAUACAGCUAAUAGGUCUCUCAUAGAAAAUUCAAUA -(((((((((((.((((((.......))))))))))))))))).----.....((((..(((...(((((.(((.....))).)))))..)))......))))................. ( -35.10) >DroSim_CAF1 2585 116 - 1 GGGCGGUCGAGCCGUAGUUAAGUACCGACUACGCUUGACCGCCC----GAAACGGGAAAGGUGAAUUUAAUAACGUCUAGGUUUUAUACAGCUAAUAGGUCUCUCAUAGAAAAUUCAAUA ((((((((((((.((((((.......))))))))))))))))))----.............(((((((.......((((((((......))))..))))(((.....))))))))))... ( -37.70) >DroEre_CAF1 1563 114 - 1 -GACGGUCGAGCCGUAGUUAAGAACCGACUGCGAUCGACCGCCU-----GAGCGGUAAAGGUGAAUUUAACAAUAUCAAGUCUGCAUACAGCUGAUAGGUCUCUCAUAGAUAAUUCAUUA -...((((((..(((((((.......))))))).))))))(((.-----....)))...((((((((......(((((.(.(((....))))))))).((((.....)))))))))))). ( -32.30) >DroYak_CAF1 2628 119 - 1 -GACGGUCGAGCCGUAGUUAAGCACCGACUACGAUCAGGCGGUCGAUCGUAGCGGAGAGGGUGAAUUUAACAAUAUCAAGUCUGCAUACAGCUGAUAGGUCUCUCAUAGAUAAUUCAUUA -.((((.....)))).........(((.((((((((........)))))))))))....((((((((......(((((.(.(((....))))))))).((((.....)))))))))))). ( -32.90) >consensus _GGCGGUCGAGCCGUAGUUAAGUACCGACUACGCUCGACCGCCC____GAAACGGGAAAGGUGAAUUUAAUAAUAUCAAGUUUGUAUACAGCUAAUAGGUCUCUCAUAGAAAAUUCAAUA .(((((((((((.((((((.......)))))))))))))))))........(((((...((((..........))))...)))))................................... (-21.18 = -21.26 + 0.08)

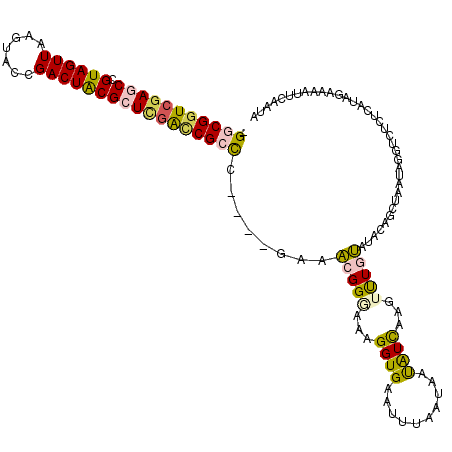
| Location | 11,408,609 – 11,408,718 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 85.54 |
| Mean single sequence MFE | -45.08 |
| Consensus MFE | -34.37 |
| Energy contribution | -36.29 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11408609 109 + 22224390 CAUAAUAUUAUUAAAUUCACCUUUCCCGU-UC----GGGCGAUCGAGCGUAGUCGGUACUUAACUACGGCUUGACCGCC-CCGGAGAUCGGGACGUGCUGGGGGAGCUACCCCCG .................(((...(((((.-((----(((((.(((((((((((..(...)..))))).)))))).))))-)....)).))))).)))..(((((.....))))). ( -43.60) >DroSec_CAF1 1698 110 + 1 CUAGAUGUUAUUAUAUUCACCUUUCCCGUUUC----GGGCGGUCAAGCGUAGUCGGUACUUAACUACGGCUCGACCGCC-CCGGGGAUCGGGACGUGCUGGGGGAGCUACCCCCG .................(((...((((((..(----((((((((.((((((((..(...)..))))).))).)))))))-)..)..).))))).)))..(((((.....))))). ( -47.70) >DroSim_CAF1 2625 111 + 1 CUAGACGUUAUUAAAUUCACCUUUCCCGUUUC----GGGCGGUCAAGCGUAGUCGGUACUUAACUACGGCUCGACCGCCCCCGGGGAUCGGGACGUGCUGGGGGAGCUACCCCCG ..(((((((..........((.((((((....----((((((((.((((((((..(...)..))))).))).)))))))).))))))..))))))).))(((((.....))))). ( -48.90) >DroEre_CAF1 1603 109 + 1 CUUGAUAUUGUUAAAUUCACCUUUACCGCUC-----AGGCGGUCGAUCGCAGUCGGUUCUUAACUACGGCUCGACCGUC-CCGGGGAUCGGGACGUGCUGGGGGAGCUACCCCCA .................(((....(((((..-----..))))).(.(((.(((((...........)))))))).)(((-(((.....)))))))))..(((((.....))))). ( -38.40) >DroYak_CAF1 2668 114 + 1 CUUGAUAUUGUUAAAUUCACCCUCUCCGCUACGAUCGACCGCCUGAUCGUAGUCGGUGCUUAACUACGGCUCGACCGUC-CCGGGGAUCGGGCCGUGCUGGGGGAGCUACCCCCG .........(((((...((((......((((((((((......)))))))))).)))).)))))((((((((((((...-....)).))))))))))..(((((.....))))). ( -46.80) >consensus CUUGAUAUUAUUAAAUUCACCUUUCCCGUUUC____GGGCGGUCGAGCGUAGUCGGUACUUAACUACGGCUCGACCGCC_CCGGGGAUCGGGACGUGCUGGGGGAGCUACCCCCG .................(((...(((((.........((((((((((((((((..(...)..))))).))))))))))).........))))).)))..(((((.....))))). (-34.37 = -36.29 + 1.92)



| Location | 11,408,609 – 11,408,718 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 85.54 |
| Mean single sequence MFE | -48.96 |
| Consensus MFE | -39.41 |
| Energy contribution | -40.65 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.80 |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.998467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11408609 109 - 22224390 CGGGGGUAGCUCCCCCAGCACGUCCCGAUCUCCGG-GGCGGUCAAGCCGUAGUUAAGUACCGACUACGCUCGAUCGCCC----GA-ACGGGAAAGGUGAAUUUAAUAAUAUUAUG .(((((.....)))))..(((.(((((.((....(-(((((((.(((.((((((.......))))))))).))))))))----))-.)))))...)))................. ( -47.70) >DroSec_CAF1 1698 110 - 1 CGGGGGUAGCUCCCCCAGCACGUCCCGAUCCCCGG-GGCGGUCGAGCCGUAGUUAAGUACCGACUACGCUUGACCGCCC----GAAACGGGAAAGGUGAAUAUAAUAACAUCUAG .(((((.....)))))..(((.(((((.......(-(((((((((((.((((((.......))))))))))))))))))----....)))))...)))................. ( -52.30) >DroSim_CAF1 2625 111 - 1 CGGGGGUAGCUCCCCCAGCACGUCCCGAUCCCCGGGGGCGGUCGAGCCGUAGUUAAGUACCGACUACGCUUGACCGCCC----GAAACGGGAAAGGUGAAUUUAAUAACGUCUAG .(((((.....)))))..(((.(((((..(....)((((((((((((.((((((.......))))))))))))))))))----....)))))...)))................. ( -53.80) >DroEre_CAF1 1603 109 - 1 UGGGGGUAGCUCCCCCAGCACGUCCCGAUCCCCGG-GACGGUCGAGCCGUAGUUAAGAACCGACUGCGAUCGACCGCCU-----GAGCGGUAAAGGUGAAUUUAACAAUAUCAAG ((((((.....))))))((..((((((.....)))-)))((((((..(((((((.......))))))).))))))....-----..))((((...((.......))..))))... ( -46.80) >DroYak_CAF1 2668 114 - 1 CGGGGGUAGCUCCCCCAGCACGGCCCGAUCCCCGG-GACGGUCGAGCCGUAGUUAAGCACCGACUACGAUCAGGCGGUCGAUCGUAGCGGAGAGGGUGAAUUUAACAAUAUCAAG .(((((.....)))))((((((((.(((((.....-...))))).))))).))).....(((.((((((((........))))))))))).((..((.......))....))... ( -44.20) >consensus CGGGGGUAGCUCCCCCAGCACGUCCCGAUCCCCGG_GGCGGUCGAGCCGUAGUUAAGUACCGACUACGCUCGACCGCCC____GAAACGGGAAAGGUGAAUUUAAUAAUAUCAAG .(((((.....)))))..(((.(((((.........(((((((((((.((((((.......))))))))))))))))).........)))))...)))................. (-39.41 = -40.65 + 1.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:31 2006