
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,403,176 – 11,403,311 |
| Length | 135 |
| Max. P | 0.999491 |

| Location | 11,403,176 – 11,403,271 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.37 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -18.30 |
| Energy contribution | -18.67 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.65 |
| SVM decision value | 3.65 |
| SVM RNA-class probability | 0.999491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
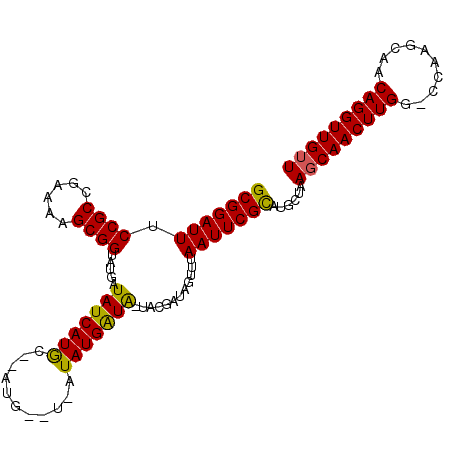
>X_DroMel_CAF1 11403176 95 + 22224390 UCGGAAUUAAACAAAAUACAUUCCGAAUAUUGAUUAGAUGGCGGAUUUCCGCCGAAAAGCGGUAUGAUAUCAUGC---------CUAUGAUG----------------UACGAUAGUUUA (((((((............))))))).(((((......((((((....))))))......((((((....)))))---------).......----------------..)))))..... ( -27.80) >DroSec_CAF1 4485 104 + 1 UCGGAAUUAAACAAAAUACAGUCCGAAUAUUGAUUAGACGGCGGAUUUCCGCCGAAGAGCGGAAUGAUAUCAUAGGCAUGAAUGAUAUGAUA----------------UACGAUAGUUUA (((((................)))))............((((((....))))))..((((......((((((((..((....)).)))))))----------------)......)))). ( -26.69) >DroEre_CAF1 2999 118 + 1 UCAAAAUAAGGCUGAAUACAAUCCGAAUAUUGAUUAGGCGGCGGAUUUCCGCCGAAGUG-GGUAUGAUAUCAUGUCUAUAG-UAUUAUGGUAUUAUGGUAAAUGUAUGUACGAAAGUGUA ...............(((((..(((((((((.......((((((....))))))..(((-(..(((....)))..))))..-......)))))).)))....))))).(((....))).. ( -30.00) >DroYak_CAF1 1940 103 + 1 UCAAAAUAAAACAAAAUACAUUCCGAAUAUUGAUUAGGCGGCGGAUUUCCGCUGAAAAGCGGUAUGAUAUCAUGCCUAUGG-UAUUAUGGUAU----------------GCGAAAGUGUA ......................(((((((((...((((((....(((.(((((....)))))...)))....)))))).))-)))).)))(((----------------((....))))) ( -27.80) >consensus UCAAAAUAAAACAAAAUACAUUCCGAAUAUUGAUUAGACGGCGGAUUUCCGCCGAAAAGCGGUAUGAUAUCAUGCCUAUGG_UAUUAUGAUA________________UACGAAAGUGUA ...............((((.(((...((((((......((((((....)))))).....))))))..(((((((...........)))))))...................))).)))). (-18.30 = -18.67 + 0.38)



| Location | 11,403,176 – 11,403,271 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.37 |
| Mean single sequence MFE | -19.89 |
| Consensus MFE | -12.07 |
| Energy contribution | -12.32 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11403176 95 - 22224390 UAAACUAUCGUA----------------CAUCAUAG---------GCAUGAUAUCAUACCGCUUUUCGGCGGAAAUCCGCCAUCUAAUCAAUAUUCGGAAUGUAUUUUGUUUAAUUCCGA ............----------------.......(---------(.(((....))).)).......(((((....))))).............(((((((............))))))) ( -21.60) >DroSec_CAF1 4485 104 - 1 UAAACUAUCGUA----------------UAUCAUAUCAUUCAUGCCUAUGAUAUCAUUCCGCUCUUCGGCGGAAAUCCGCCGUCUAAUCAAUAUUCGGACUGUAUUUUGUUUAAUUCCGA (((((....(((----------------((..(((((((........)))))))...((((.....((((((....)))))).............)))).)))))...)))))....... ( -21.97) >DroEre_CAF1 2999 118 - 1 UACACUUUCGUACAUACAUUUACCAUAAUACCAUAAUA-CUAUAGACAUGAUAUCAUACC-CACUUCGGCGGAAAUCCGCCGCCUAAUCAAUAUUCGGAUUGUAUUCAGCCUUAUUUUGA ......................................-...(((..(((....)))...-.....((((((....)))))).))).((((.((..((.(((....)))))..)).)))) ( -17.00) >DroYak_CAF1 1940 103 - 1 UACACUUUCGC----------------AUACCAUAAUA-CCAUAGGCAUGAUAUCAUACCGCUUUUCAGCGGAAAUCCGCCGCCUAAUCAAUAUUCGGAAUGUAUUUUGUUUUAUUUUGA .........((----------------((.((......-...((((((((....))).(((((....))))).........)))))..........)).))))................. ( -19.01) >consensus UAAACUAUCGUA________________UACCAUAAUA_CCAUAGGCAUGAUAUCAUACCGCUCUUCGGCGGAAAUCCGCCGCCUAAUCAAUAUUCGGAAUGUAUUUUGUUUAAUUCCGA ...............................................(((....)))..........(((((....))))).............(((((((............))))))) (-12.07 = -12.32 + 0.25)



| Location | 11,403,216 – 11,403,311 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.91 |
| Mean single sequence MFE | -31.43 |
| Consensus MFE | -23.51 |
| Energy contribution | -22.97 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11403216 95 + 22224390 GCGGAUUUCCGCCGAAAAGCGGUAUGAUAUCAUGC---------CUAUGAUG-UACGAUAGUUUAAUUCGCAUGCCAAGCAACUUGGCCCAAGCAACAGGUUGUU ((((....))))........(((((((((((((..---------..))))))-).(((.........))))))))).(((((((((((....))..))))))))) ( -28.70) >DroSec_CAF1 4525 104 + 1 GCGGAUUUCCGCCGAAGAGCGGAAUGAUAUCAUAGGCAUGAAUGAUAUGAUA-UACGAUAGUUUAAUUCGUAUGCUAAGCAACUUGGGCCAAGCAACAGGUUGUU ((((....)))).....(((...((.(((((((........))))))).))(-(((((.........))))))))).(((((((((.((...))..))))))))) ( -29.00) >DroYak_CAF1 1980 98 + 1 GCGGAUUUCCGCUGAAAAGCGGUAUGAUAUCAUGCCUAUGG-UAUUAUGGUAU-GCGAAAGUGUAAUUCGCAUGCUAACCAACUUGG-----GCAACAGGUUGUU ........(((((....)))))..(((((((((....))))-)))))((((((-(((((.......)))))))))))..(((((((.-----....))))))).. ( -36.60) >consensus GCGGAUUUCCGCCGAAAAGCGGUAUGAUAUCAUGC__AUG__U_AUAUGAUA_UACGAUAGUUUAAUUCGCAUGCUAAGCAACUUGG_CCAAGCAACAGGUUGUU (((((((.((((......)))).....(((((((...........)))))))............)))))))......(((((((((..........))))))))) (-23.51 = -22.97 + -0.55)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:22 2006