| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,401,389 – 11,401,501 |
| Length | 112 |
| Max. P | 0.813488 |

| Location | 11,401,389 – 11,401,501 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.58 |
| Mean single sequence MFE | -24.66 |
| Consensus MFE | -14.51 |
| Energy contribution | -14.81 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11401389 112 - 22224390 CAUUACCCGCA---CCACCACCAGCACCACCAACACCUCCUACAUUGUCCUGAACACGGGCUAUCUGAAGACUUUCCCCGGCAUCCUCAAGCUCUUCGAGCUGGUGCGUUAAUGC----- (((((..((((---((.......((....................(((.....)))((((...((....)).....)))))).......(((((...))))))))))).))))).----- ( -24.90) >DroVir_CAF1 734 112 - 1 CAUAACACGCA---CCACCACCAGCACCACCAACACCUCCUACAUUGUGCUCAACACGGGCUACAUCAAGACCUUUCCGGGUAUACUCAAGCUCUUCCAGCUGGUAAGUGACGUC----- ......(((((---(....((((((.....................(((.....)))(((((........((((....)))).......))))).....))))))..))).))).----- ( -25.56) >DroPse_CAF1 848 115 - 1 CAUUACACGUACCACCACCACCAGCACCACCAACACCUCGUACAUCGUGCUCAACACAGGCUACCUGAAGUCCUUUCCAGGAAUUCUAAAGCUCUUUGAGCUGGUAAGUUAUGCC----- ........(((..((....((((((..............((((...))))....((.(((((....(((.((((....)))).)))...))).)).)).))))))..))..))).----- ( -22.80) >DroGri_CAF1 599 111 - 1 CAUAACACGCA---CCACCACCAGCACCACCAACACCUCCUACAUUGUGCUCAAUACGGGCUACAUCAAGACCUUUCCCGGCAUUCUUAAGCUCUUCCAGCUGGUAAGUG-CGAC----- .......((((---(....((((((....................(((((((.....)))).)))..((((((......))...))))...........))))))..)))-))..----- ( -25.70) >DroWil_CAF1 256 117 - 1 CAUAACCCGUA---CCACGACCAGCACUACGAAUACCUCGUACAUAGUCCUCAAUACGGGCUAUCUAAAGACCUUUCCCGGCAUACUCAAGUUCUUUGAGCUGGUAAGUACAUAGUAUAU ........(((---(....((((((..(((((.....))))).(((((((.......))))))).((((((.(((.............))).)))))).))))))..))))......... ( -28.52) >DroMoj_CAF1 605 112 - 1 CAUAACACGCA---CUACCACCAGCACCACCAACACCUCCUACAUUGUGCUAAACACUGGCUACAUUAAGACCUUUCCGGGUAUUUUGAAGCUCUUCCAGCUGGUAAUUAAAUUG----- ...........---.....((((((.................((.(((.....))).))(((.((.....((((....))))....)).))).......))))))..........----- ( -20.50) >consensus CAUAACACGCA___CCACCACCAGCACCACCAACACCUCCUACAUUGUGCUCAACACGGGCUACAUCAAGACCUUUCCCGGCAUUCUCAAGCUCUUCCAGCUGGUAAGUAAAGAC_____ ...................((((((.....................(((.....)))(((((.........((......))........))))).....))))))............... (-14.51 = -14.81 + 0.31)

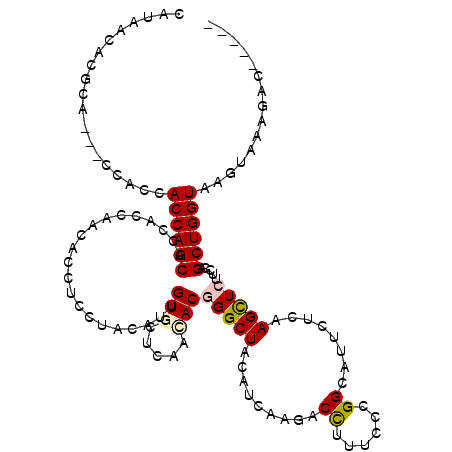

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:16 2006