
| Sequence ID | X_DroMel_CAF1 |
|---|---|
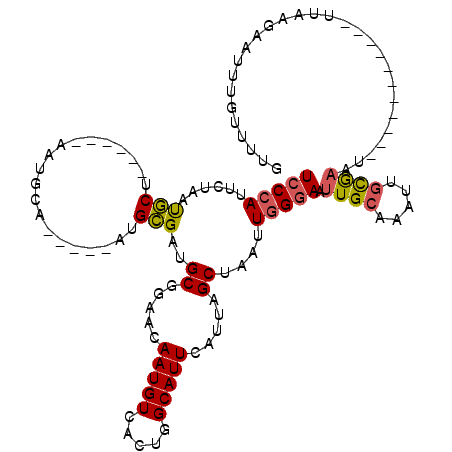
| Location | 11,391,119 – 11,391,249 |
| Length | 130 |
| Max. P | 0.977278 |

| Location | 11,391,119 – 11,391,217 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.78 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -10.33 |
| Energy contribution | -10.33 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11391119 98 - 22224390 UCCCAUUCUAAUGCU------AAUGCA-----AUGCGAUGCGGAACAAUGUCACUGGCAUUCAUUAGCUAAUUGGGAAUUGCAAAUUGCAAAU-----------UUAAGAAUUUGUUUUG (((((.......(((------((((.(-----((((.(((((......)).)).).))))))))))))....)))))....((((..((((((-----------(....))))))))))) ( -25.50) >DroSec_CAF1 44520 98 - 1 UCCCAUUCUAAUGCU------GAUGCA-----AUGCGAUGCGGAACAAUGUCACUGGCAUUCAUUAGCUAAUUGGGAAUUGCAAAUUGCGAAU-----------UUAAGAAUUUGUUUUG (((((.......(((------((((.(-----((((.(((((......)).)).).))))))))))))....)))))...(((((((.(....-----------....)))))))).... ( -25.40) >DroEre_CAF1 50700 98 - 1 UCCCAUUCUAAUGCU------AAUGCG-----AUGCGAUGCGGAACAAUGUCACUGGCAUUCAUUAGCUAAUUGGGAAUUGCAAAUUGCGAAU-----------UUAAGAAUUUGUUUUG (((((.......(((------((((.(-----((((.(((((......)).)).).))))))))))))....)))))...(((((((.(....-----------....)))))))).... ( -24.80) >DroYak_CAF1 49900 109 - 1 UCCCAUUCGAAUGCUAAUGCUAAUGCGAUGCAAUGCGAUGCGGAACAAUGUCACCAGCAUUCAUUAGCUAAUUGGGAAUUGCAAAUUGUGAAU-----------UUAAGAAUUUGUUUUG (((((.......(((((((..(((((...(((......)))((..........)).))))))))))))....)))))...(((((((.(....-----------...).))))))).... ( -26.50) >DroAna_CAF1 32305 104 - 1 UCCCAUUCCGACUCU------AA----------UGGGACGCCGAACAAUGUCACCCGCAUUUGUGAGCUAAUUUGCAAUUGCAAGCAUCGCGUUCGCGUUCGCAUUCGGAUUCUACAUCU ......(((((....------.(----------((((((((.(((((((((.....))))).(((((((....(((....)))))).)))))))))))))).)))))))).......... ( -30.70) >consensus UCCCAUUCUAAUGCU______AAUGCA_____AUGCGAUGCGGAACAAUGUCACUGGCAUUCAUUAGCUAAUUGGGAAUUGCAAAUUGCGAAU___________UUAAGAAUUUGUUUUG (((((......(((....................)))..((.....(((((.....))))).....))....))))).((((.....))))............................. (-10.33 = -10.33 + 0.00)


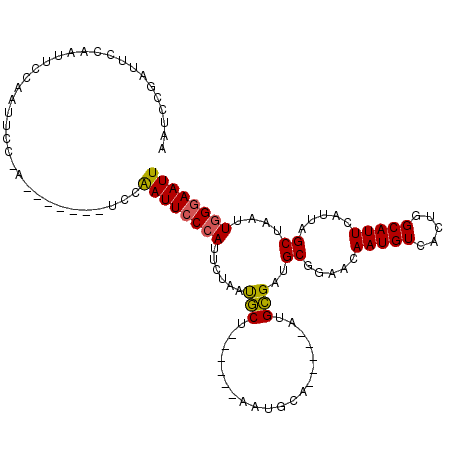
| Location | 11,391,148 – 11,391,249 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 73.60 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -12.01 |
| Energy contribution | -11.77 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11391148 101 - 22224390 AAUCCGAUUUCAAAUUCAAUUCCAAU------UCCAAUUCCCAUUCUAAUGCU------AAUGCA-----AUGCGAUGCGGAACAAUGUCACUGGCAUUCAUUAGCUAAUUGGGAAUU ..........................------...((((((((.......(((------((((.(-----((((.(((((......)).)).).))))))))))))....)))))))) ( -24.30) >DroSec_CAF1 44549 107 - 1 AAUCCGAUUUCAAUUUCAAUUCCAAUUCCGAUUCCAAUUCCCAUUCUAAUGCU------GAUGCA-----AUGCGAUGCGGAACAAUGUCACUGGCAUUCAUUAGCUAAUUGGGAAUU ...................................((((((((.......(((------((((.(-----((((.(((((......)).)).).))))))))))))....)))))))) ( -24.40) >DroEre_CAF1 50729 89 - 1 AAUCCGAUU------CCGAUUUC------------AAUUCCCAUUCUAAUGCU------AAUGCG-----AUGCGAUGCGGAACAAUGUCACUGGCAUUCAUUAGCUAAUUGGGAAUU .........------........------------((((((((.......(((------((((.(-----((((.(((((......)).)).).))))))))))))....)))))))) ( -23.80) >DroYak_CAF1 49929 106 - 1 AAUCCAAUUCCAAUUCCAAUUUC------------AAUUCCCAUUCGAAUGCUAAUGCUAAUGCGAUGCAAUGCGAUGCGGAACAAUGUCACCAGCAUUCAUUAGCUAAUUGGGAAUU .......................------------((((((((.......(((((((..(((((...(((......)))((..........)).))))))))))))....)))))))) ( -24.80) >DroAna_CAF1 32345 96 - 1 AUGCUGAUUCUGAUUCUGAUUCUGAA------UCCGAUUCCCAUUCCGACUCU------AA----------UGGGACGCCGAACAAUGUCACCCGCAUUUGUGAGCUAAUUUGCAAUU .(((.(((((.(((((.......)))------)).)).(((((((........------))----------))))).((...(((((((.....))).))))..))..))).)))... ( -19.80) >consensus AAUCCGAUUCCAAUUCCAAUUCC_A_______UCCAAUUCCCAUUCUAAUGCU______AAUGCA_____AUGCGAUGCGGAACAAUGUCACUGGCAUUCAUUAGCUAAUUGGGAAUU ...................................((((((((......(((....................)))..((.....(((((.....))))).....))....)))))))) (-12.01 = -11.77 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:08 2006