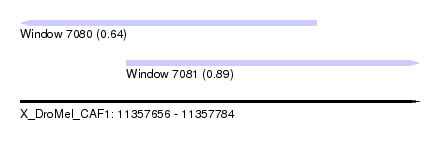
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,357,656 – 11,357,784 |
| Length | 128 |
| Max. P | 0.891193 |

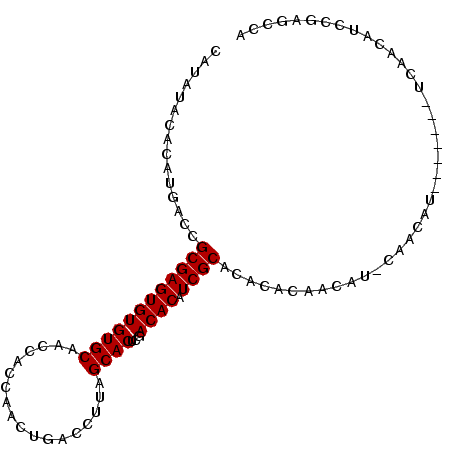
| Location | 11,357,656 – 11,357,751 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -30.95 |
| Consensus MFE | -18.04 |
| Energy contribution | -18.48 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11357656 95 - 22224390 GCGAUGUGUCGAGUGCUAAGGUCAGUUGGUGGUUGCACACACUCGCGGUCA----------------UGUGUGUAUGCAUAUGUGUGCAUAUGUGUUGCUUGCAAUUUGUU ((((((((((((.((((((......)))))).))).))))).))((((.((----------------(((((((((((....)))))))))))))))))..))........ ( -33.10) >DroSec_CAF1 18585 93 - 1 GCGAUGUGUCGAGUGCUAAGGUCAGUUGGUGGUUGCACACACUCGCGGUCA----------------UGUGUGUAUGCACAUGUGUGCA--AGUGUUGCUUGCAAUUUGUU ((((((((((((.((((((......)))))).))).))))).))))...((----------------((((((....))))))))((((--((.....))))))....... ( -34.50) >DroSim_CAF1 1179 93 - 1 GCGAUGUGUCGAGUGCUAAGGUCAGUUGGUGGUUGCACACACUCGCGGUCA----------------UGUGUAUAUGCACAUGUGUGCA--UGUGUUGCUUGCAAUUUGUU ((((((((((((.((((((......)))))).))).))))).))))...((----------------..((((...(((((((....))--)))))....))))...)).. ( -31.00) >DroEre_CAF1 22240 87 - 1 GCGAUGUGUCGAGUGCUAAGGUCAGUUGGUGGUUGCACACACUCGCGGUCA----------------UGUGUAUAUGCAUGUGU--------GUGUUACUUGCAAUUUGUU ((((((((((((.((((((......)))))).))).))))).)))).....----------------........((((.(((.--------....))).))))....... ( -24.30) >DroYak_CAF1 23556 87 - 1 GCGAUGUGUCGAGUGCUAAGGUCAGUUGGUGGUUGCACACACUCGCGGUCA----------------UGUGCAUAUGCAUGUGU--------GUGUUACUUGCAAUUUGUU ((((((((((((.((((((......)))))).))).))))).))))...((----------------(((((....))))))).--------.(((.....)))....... ( -25.80) >DroAna_CAF1 13128 101 - 1 ---GUCUGUCGAGAGCUAAGGUCAGCUAGUGGUUGCACACACGCUCGCACACACUCACCGAGUUCCGAGUGCCUGUGGGCCUGUGU------GCGUUUC-UGCAAUUUGUU ---..........((((......)))).(..(.((((((((.(((((((..(((((..........)))))..))))))).)))))------)))...)-..)........ ( -37.00) >consensus GCGAUGUGUCGAGUGCUAAGGUCAGUUGGUGGUUGCACACACUCGCGGUCA________________UGUGUAUAUGCACAUGUGU______GUGUUGCUUGCAAUUUGUU ((((((((((((.((((((......)))))).))).))))).)))).......................((((...((((............))))....))))....... (-18.04 = -18.48 + 0.45)

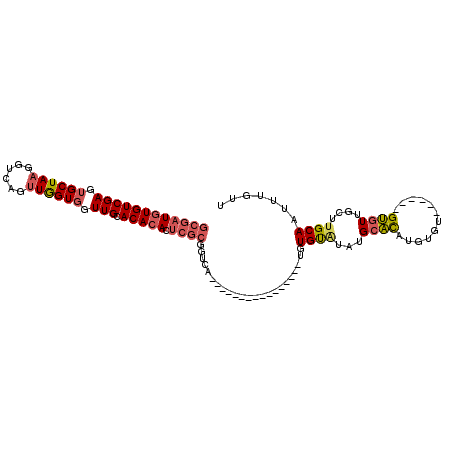
| Location | 11,357,690 – 11,357,784 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -13.63 |
| Consensus MFE | -13.37 |
| Energy contribution | -13.37 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.891193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11357690 94 + 22224390 CAUACACACAUGACCGCGAGUGUGUGCAACCACCAACUGACCUUAGCACUCGACACAUCGCACACACAACAUUCAACAU-------UCAACAUCCGAGCCA ...............((..((((((((.........(((....))).....((....))))))))))............-------((.......)))).. ( -13.50) >DroSec_CAF1 18617 94 + 1 CAUACACACAUGACCGCGAGUGUGUGCAACCACCAACUGACCUUAGCACUCGACACAUCGCACACACAACAUCCAACAU-------UCAACAUCCGAGCCA ...............((..((((((((.........(((....))).....((....))))))))))............-------((.......)))).. ( -13.50) >DroSim_CAF1 1211 101 + 1 CAUAUACACAUGACCGCGAGUGUGUGCAACCACCAACUGACCUUAGCACUCGACACAUCGCACACACAACAUCCAACAUUCAACAUUCAACAUCCGAGCCA ...............((..((((((((.........(((....))).....((....))))))))))...................((.......)))).. ( -13.50) >DroEre_CAF1 22266 84 + 1 CAUAUACACAUGACCGCGAGUGUGUGCAACCACCAACUGACCUUAGCACUCGACACAUCGCACAC---------AACAU-------CCAACAUCCGAG-CA ...............((((((((((((..................))))...)))).))))....---------.....-------............-.. ( -13.37) >DroYak_CAF1 23582 84 + 1 CAUAUGCACAUGACCGCGAGUGUGUGCAACCACCAACUGACCUUAGCACUCGACACAUCGCACAC---------AACAU-------CCAACAUCCGAG-CA ....(((........((((((((((((..................))))...)))).))))....---------....(-------(........)))-)) ( -14.27) >consensus CAUAUACACAUGACCGCGAGUGUGUGCAACCACCAACUGACCUUAGCACUCGACACAUCGCACACACAACAU_CAACAU_______UCAACAUCCGAGCCA ...............((((((((((((..................))))...)))).))))........................................ (-13.37 = -13.37 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:46 2006